Custom Sampling Routines in StochTree
CustomSamplingRoutine.RmdMotivation
While the functions bart() and bcf()
provide simple and performant interfaces for supervised learning /
causal inference, stochtree also offers access to many of
the “low-level” data structures that are typically implemented in C++.
This low-level interface is not designed for performance or even
simplicity — rather the intent is to provide a “prototype” interface to
the C++ code that doesn’t require modifying any C++.
To illustrate when such a prototype interface might be useful, consider the classic BART algorithm:
OUTPUT: samples of a decision forest with trees and global variance parameter
Initialize via a default or a data-dependent calibration exercise
Initialize “forest 0” with trees with a single root node, referring to tree ’s prediction vector as
Compute residual as
FOR IN :
Initialize forest from forest
FOR IN :
Add predictions for tree to residual:
Update tree via Metropolis-Hastings with and as data and tree priors depending on (, , , )
Sample leaf node parameters for tree via Gibbs (leaf node prior is )
Subtract (updated) predictions for tree from residual:
Sample via Gibbs (prior is )
While the algorithm itself is conceptually simple, much of the core computation is carried out in low-level languages such as C or C++ because of the tree data structure. As a result, any changes to this algorithm, such as supporting heteroskedasticity (Pratola et al. (2020)), categorical outcomes (Murray (2021)) or causal effect estimation (Hahn, Murray, and Carvalho (2020)) require modifying low-level code.
The prototype interface exposes the core components of the loop above at the R level, thus making it possible to interchange C++ computation for steps like “update tree via Metropolis-Hastings” with R computation for a custom variance model, other user-specified additive mean model components, and so on.
To begin, load the stochtree package
Demo 1: Supervised Learning
Simulation
Simulate a simple partitioned linear model
# Generate the data
n <- 500
p_X <- 10
p_W <- 1
X <- matrix(runif(n*p_X), ncol = p_X)
W <- matrix(runif(n*p_W), ncol = p_W)
f_XW <- (
((0 <= X[,1]) & (0.25 > X[,1])) * (-3*W[,1]) +
((0.25 <= X[,1]) & (0.5 > X[,1])) * (-1*W[,1]) +
((0.5 <= X[,1]) & (0.75 > X[,1])) * (1*W[,1]) +
((0.75 <= X[,1]) & (1 > X[,1])) * (3*W[,1])
)
y <- f_XW + rnorm(n, 0, 1)
# Standardize outcome
y_bar <- mean(y)
y_std <- sd(y)
resid <- (y-y_bar)/y_stdSampling
Set some parameters that inform the forest and variance parameter samplers
alpha <- 0.9
beta <- 1.25
min_samples_leaf <- 1
max_depth <- 10
num_trees <- 100
cutpoint_grid_size = 100
global_variance_init = 1.
tau_init = 0.5
leaf_prior_scale = matrix(c(tau_init), ncol = 1)
nu <- 4
lambda <- 0.5
a_leaf <- 2.
b_leaf <- 0.5
leaf_regression <- T
feature_types <- as.integer(rep(0, p_X)) # 0 = numeric
var_weights <- rep(1/p_X, p_X)Initialize R-level access to the C++ classes needed to sample our model
# Data
if (leaf_regression) {
forest_dataset <- createForestDataset(X, W)
outcome_model_type <- 1
} else {
forest_dataset <- createForestDataset(X)
outcome_model_type <- 0
}
outcome <- createOutcome(resid)
# Random number generator (std::mt19937)
rng <- createRNG()
# Sampling data structures
forest_model <- createForestModel(forest_dataset, feature_types,
num_trees, n, alpha, beta,
min_samples_leaf, max_depth)
# "Active forest" (which gets updated by the sample) and
# container of forest samples (which is written to when
# a sample is not discarded due to burn-in / thinning)
if (leaf_regression) {
forest_samples <- createForestContainer(num_trees, 1, F)
active_forest <- createForest(num_trees, 1, F)
} else {
forest_samples <- createForestContainer(num_trees, 1, T)
active_forest <- createForest(num_trees, 1, T)
}Prepare to run the sampler
num_warmstart <- 10
num_mcmc <- 100
num_samples <- num_warmstart + num_mcmc
global_var_samples <- c(global_variance_init, rep(0, num_samples))
leaf_scale_samples <- c(tau_init, rep(0, num_samples))Run the grow-from-root sampler to “warm-start” BART
for (i in 1:num_warmstart) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = T
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
}Pick up from the last GFR forest (and associated global variance / leaf scale parameters) with an MCMC sampler
for (i in (num_warmstart+1):num_samples) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = F
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
}Predict and rescale samples
# Forest predictions
preds <- forest_samples$predict(forest_dataset)*y_std + y_bar
# Global error variance
sigma_samples <- sqrt(global_var_samples)*y_stdResults
Inspect the initial samples obtained via “grow-from-root” (He and Hahn (2023))
plot(sigma_samples[1:num_warmstart], ylab="sigma")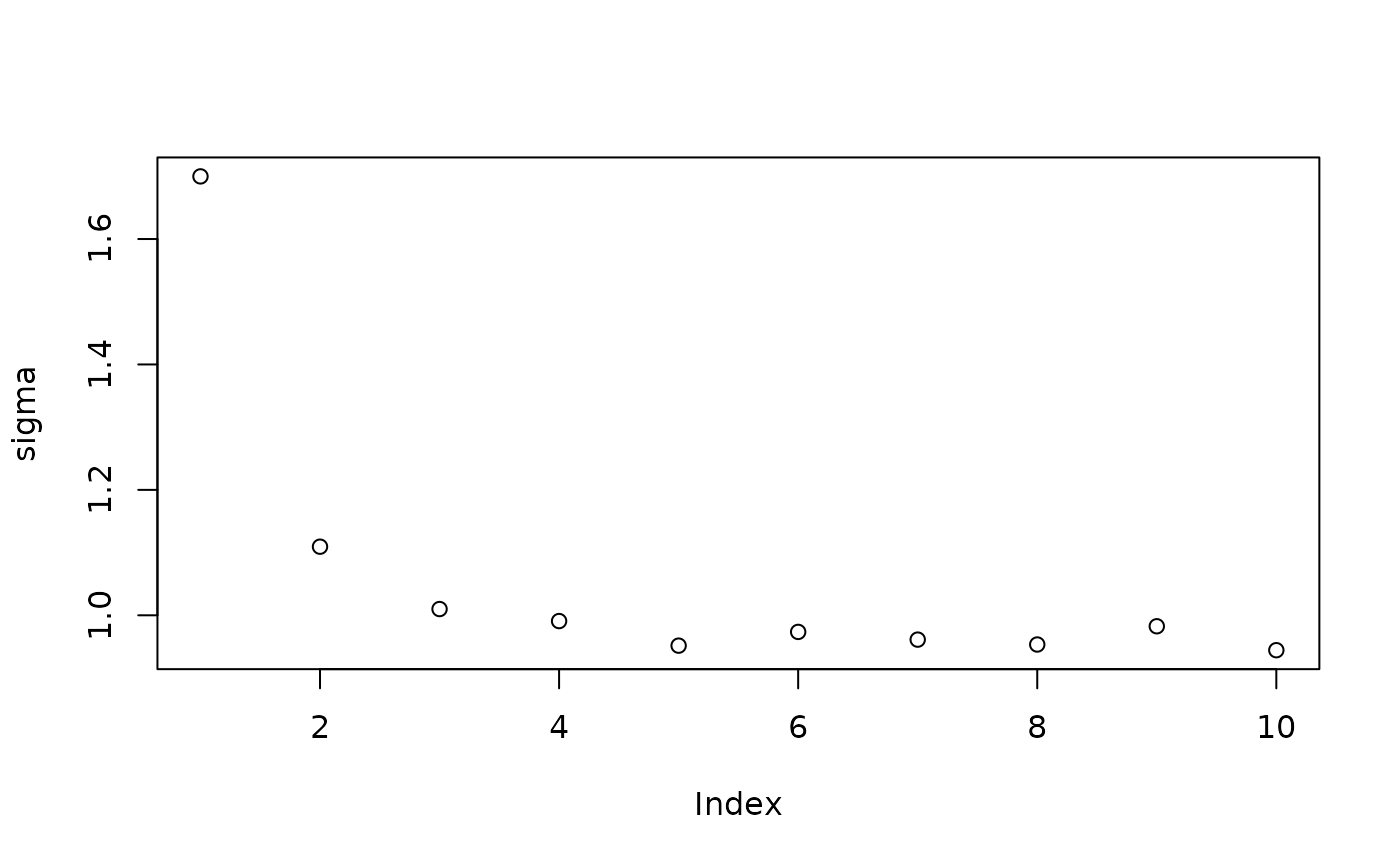
plot(rowMeans(preds[,1:num_warmstart]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)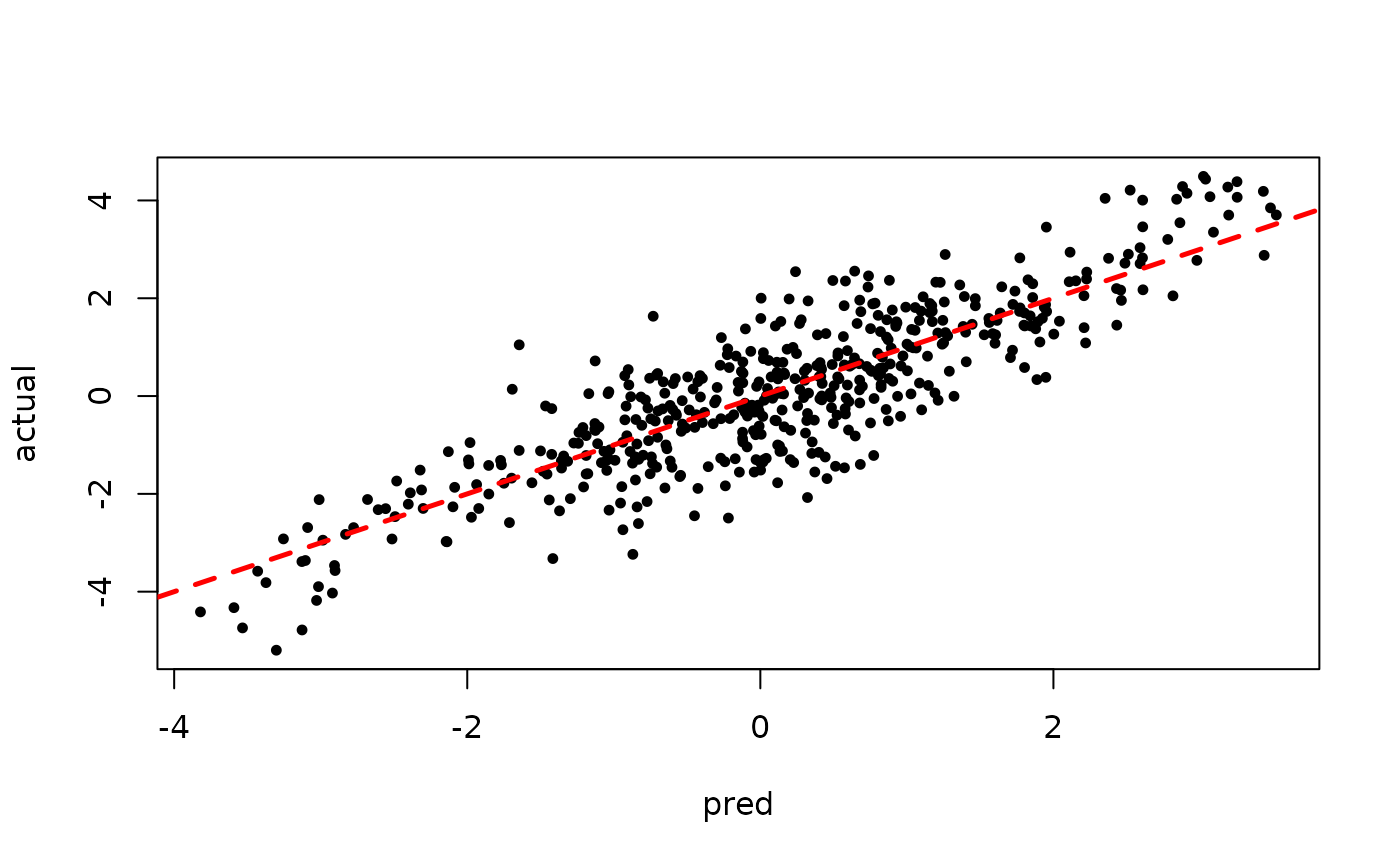
Inspect the BART samples obtained after “warm-starting”
plot(sigma_samples[(num_warmstart+1):num_samples], ylab="sigma")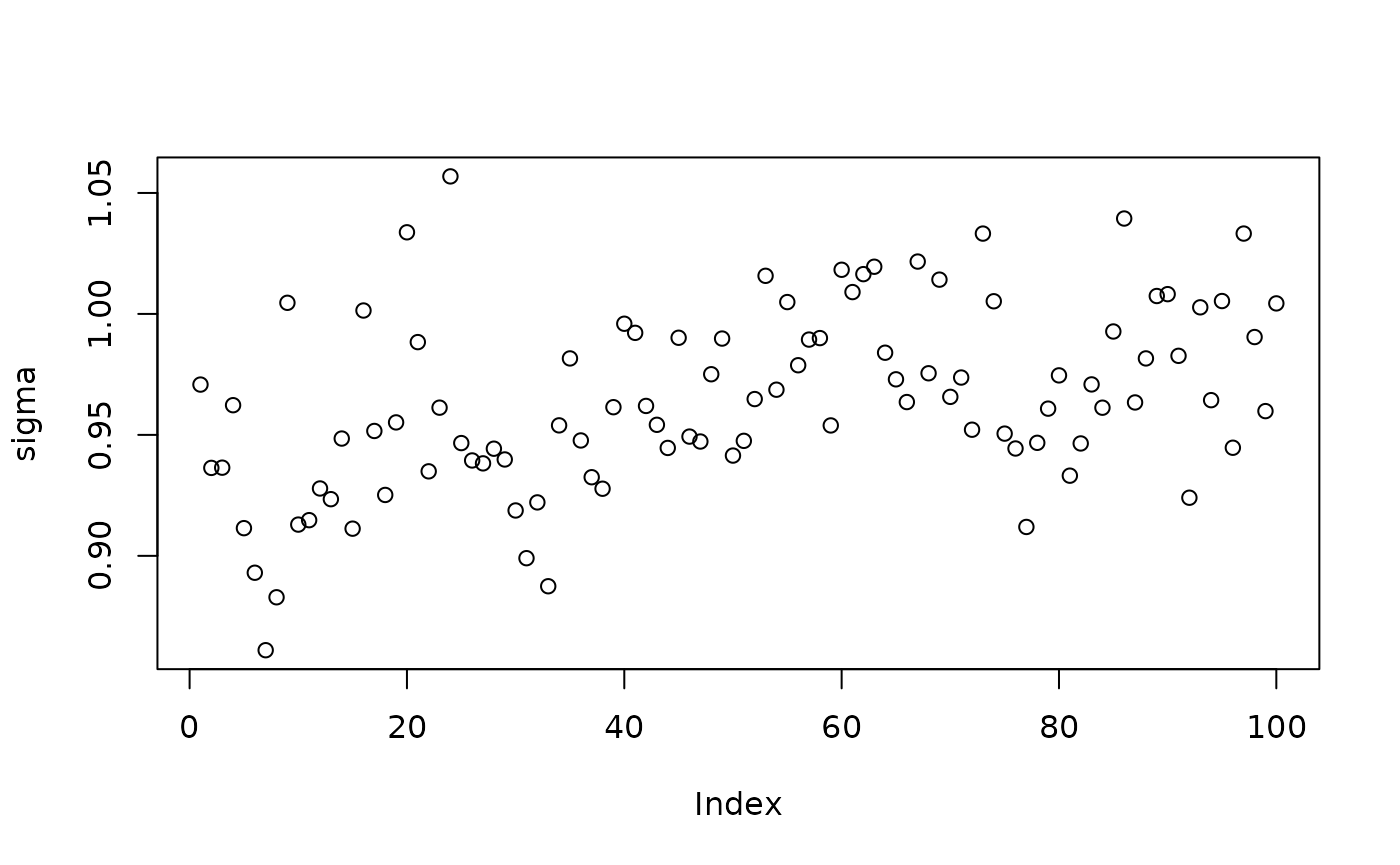
plot(rowMeans(preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)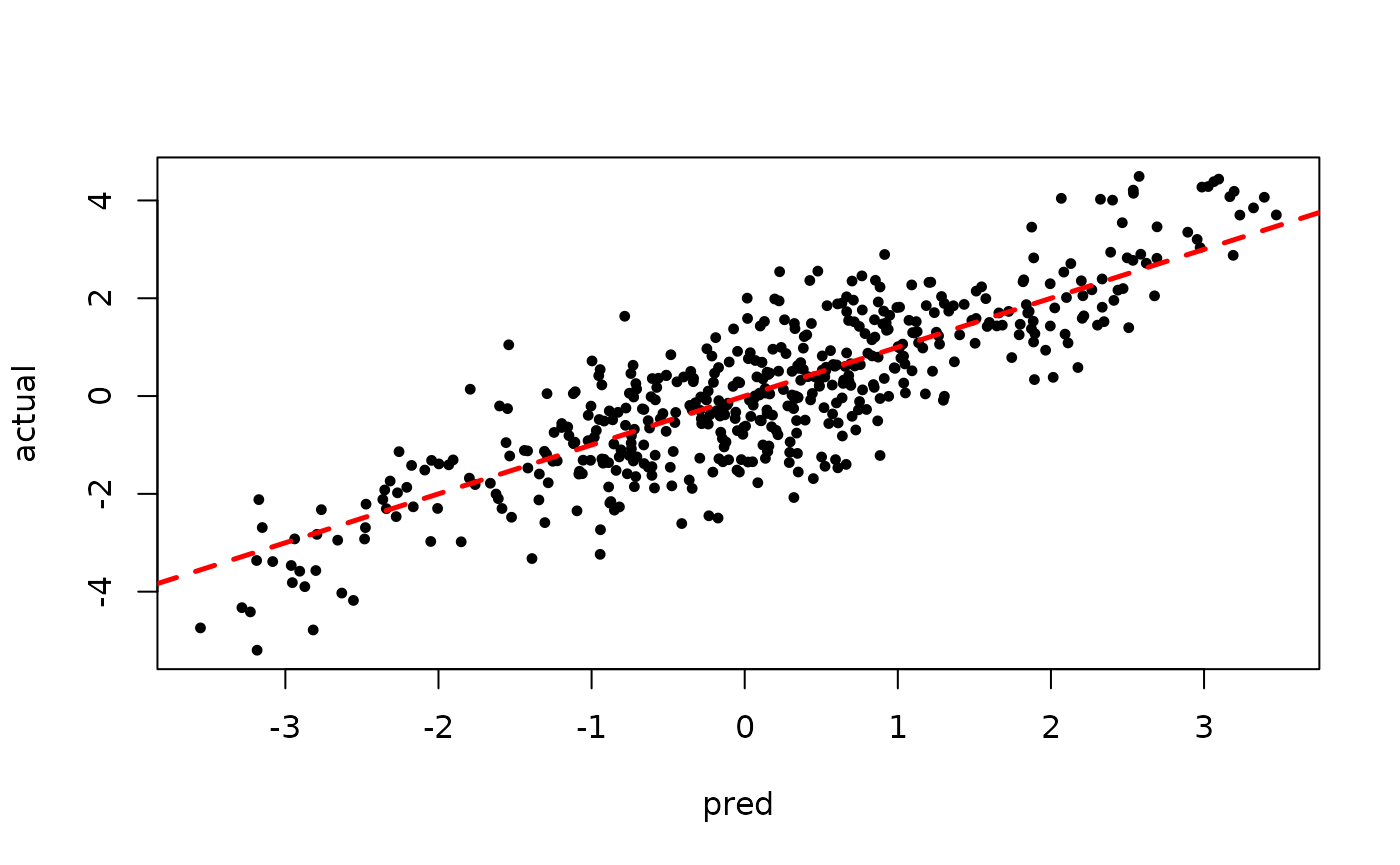
Demo 2: Supervised Learning with Additive Random Effects
We build on the above example and add a simple “random effects” structure: every observation is in either group 1 or group 2 and there is a random group intercept (simulated to be quite strong, underscoring the need for random effects modeling).
Simulation
Simulate a partitioned linear model with a simple additive group random effect structure
# Generate the data
n <- 500
p_X <- 10
p_W <- 1
X <- matrix(runif(n*p_X), ncol = p_X)
W <- matrix(runif(n*p_W), ncol = p_W)
group_ids <- rep(c(1,2), n %/% 2)
rfx_coefs <- c(-5, 5)
rfx_basis <- rep(1, n)
f_XW <- (
((0 <= X[,1]) & (0.25 > X[,1])) * (-3*W[,1]) +
((0.25 <= X[,1]) & (0.5 > X[,1])) * (-1*W[,1]) +
((0.5 <= X[,1]) & (0.75 > X[,1])) * (1*W[,1]) +
((0.75 <= X[,1]) & (1 > X[,1])) * (3*W[,1])
)
rfx_term <- rfx_coefs[group_ids] * rfx_basis
y <- f_XW + rfx_term + rnorm(n, 0, 1)
# Standardize outcome
y_bar <- mean(y)
y_std <- sd(y)
resid <- (y-y_bar)/y_stdSampling
Set some parameters that inform the forest and variance parameter samplers
alpha <- 0.9
beta <- 1.25
min_samples_leaf <- 1
max_depth <- 10
num_trees <- 100
cutpoint_grid_size = 100
global_variance_init = 1.
tau_init = 0.5
leaf_prior_scale = matrix(c(tau_init), ncol = 1)
nu <- 4
lambda <- 0.5
a_leaf <- 2.
b_leaf <- 0.5
leaf_regression <- T
feature_types <- as.integer(rep(0, p_X)) # 0 = numeric
var_weights <- rep(1/p_X, p_X)Set some parameters that inform the random effects samplers
alpha_init <- c(1)
xi_init <- matrix(c(1,1),1,2)
sigma_alpha_init <- matrix(c(1),1,1)
sigma_xi_init <- matrix(c(1),1,1)
sigma_xi_shape <- 1
sigma_xi_scale <- 1Initialize R-level access to the C++ classes needed to sample our model
# Data
if (leaf_regression) {
forest_dataset <- createForestDataset(X, W)
outcome_model_type <- 1
} else {
forest_dataset <- createForestDataset(X)
outcome_model_type <- 0
}
outcome <- createOutcome(resid)
# Random number generator (std::mt19937)
rng <- createRNG()
# Sampling data structures
forest_model <- createForestModel(forest_dataset, feature_types,
num_trees, n, alpha, beta,
min_samples_leaf, max_depth)
# "Active forest" (which gets updated by the sample) and
# container of forest samples (which is written to when
# a sample is not discarded due to burn-in / thinning)
if (leaf_regression) {
forest_samples <- createForestContainer(num_trees, 1, F)
active_forest <- createForest(num_trees, 1, F)
} else {
forest_samples <- createForestContainer(num_trees, 1, T)
active_forest <- createForest(num_trees, 1, T)
}
# Random effects dataset
rfx_basis <- as.matrix(rfx_basis)
group_ids <- as.integer(group_ids)
rfx_dataset <- createRandomEffectsDataset(group_ids, rfx_basis)
# Random effects details
num_groups <- length(unique(group_ids))
num_components <- ncol(rfx_basis)
# Random effects tracker
rfx_tracker <- createRandomEffectsTracker(group_ids)
# Random effects model
rfx_model <- createRandomEffectsModel(num_components, num_groups)
rfx_model$set_working_parameter(alpha_init)
rfx_model$set_group_parameters(xi_init)
rfx_model$set_working_parameter_cov(sigma_alpha_init)
rfx_model$set_group_parameter_cov(sigma_xi_init)
rfx_model$set_variance_prior_shape(sigma_xi_shape)
rfx_model$set_variance_prior_scale(sigma_xi_scale)
# Random effect samples
rfx_samples <- createRandomEffectSamples(num_components, num_groups, rfx_tracker)Prepare to run the sampler
num_warmstart <- 10
num_mcmc <- 100
num_samples <- num_warmstart + num_mcmc
global_var_samples <- c(global_variance_init, rep(0, num_samples))
leaf_scale_samples <- c(tau_init, rep(0, num_samples))Run the grow-from-root sampler to “warm-start” BART
for (i in 1:num_warmstart) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = T
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
# Sample random effects model
rfx_model$sample_random_effect(rfx_dataset, outcome, rfx_tracker, rfx_samples,
TRUE, global_var_samples[i+1], rng)
}Pick up from the last GFR forest (and associated global variance / leaf scale parameters) with an MCMC sampler
for (i in (num_warmstart+1):num_samples) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = F
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
# Sample random effects model
rfx_model$sample_random_effect(rfx_dataset, outcome, rfx_tracker, rfx_samples,
TRUE, global_var_samples[i+1], rng)
}Predict and rescale samples
# Forest predictions
forest_preds <- forest_samples$predict(forest_dataset)*y_std + y_bar
# Random effects predictions
rfx_preds <- rfx_samples$predict(group_ids, rfx_basis)*y_std
# Overall predictions
preds <- forest_preds + rfx_preds
# Global error variance
sigma_samples <- sqrt(global_var_samples)*y_stdResults
Inspect the initial samples obtained via grow-from-root and an additive random effects model
plot(sigma_samples[1:num_warmstart], ylab="sigma")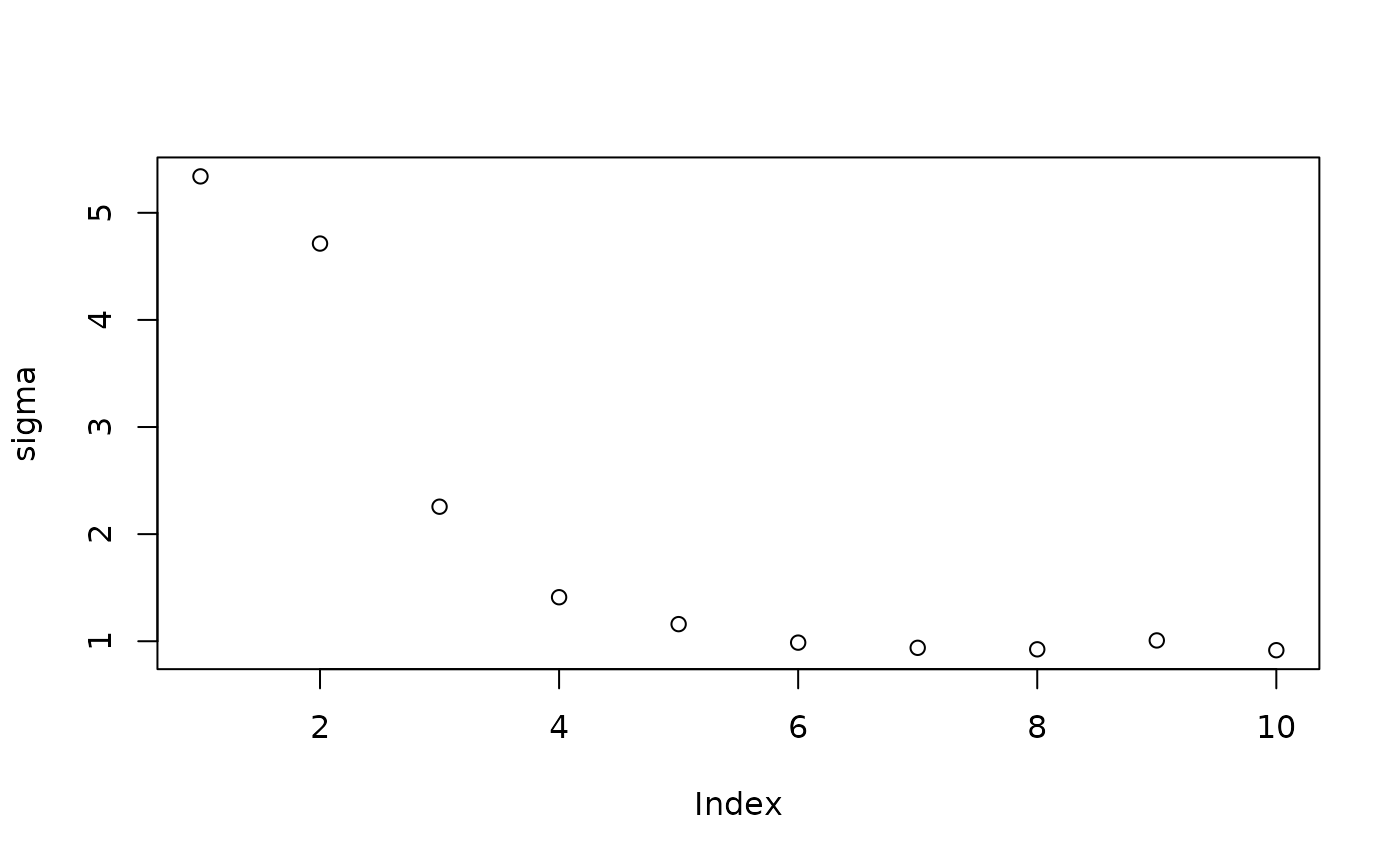
plot(rowMeans(preds[,1:num_warmstart]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)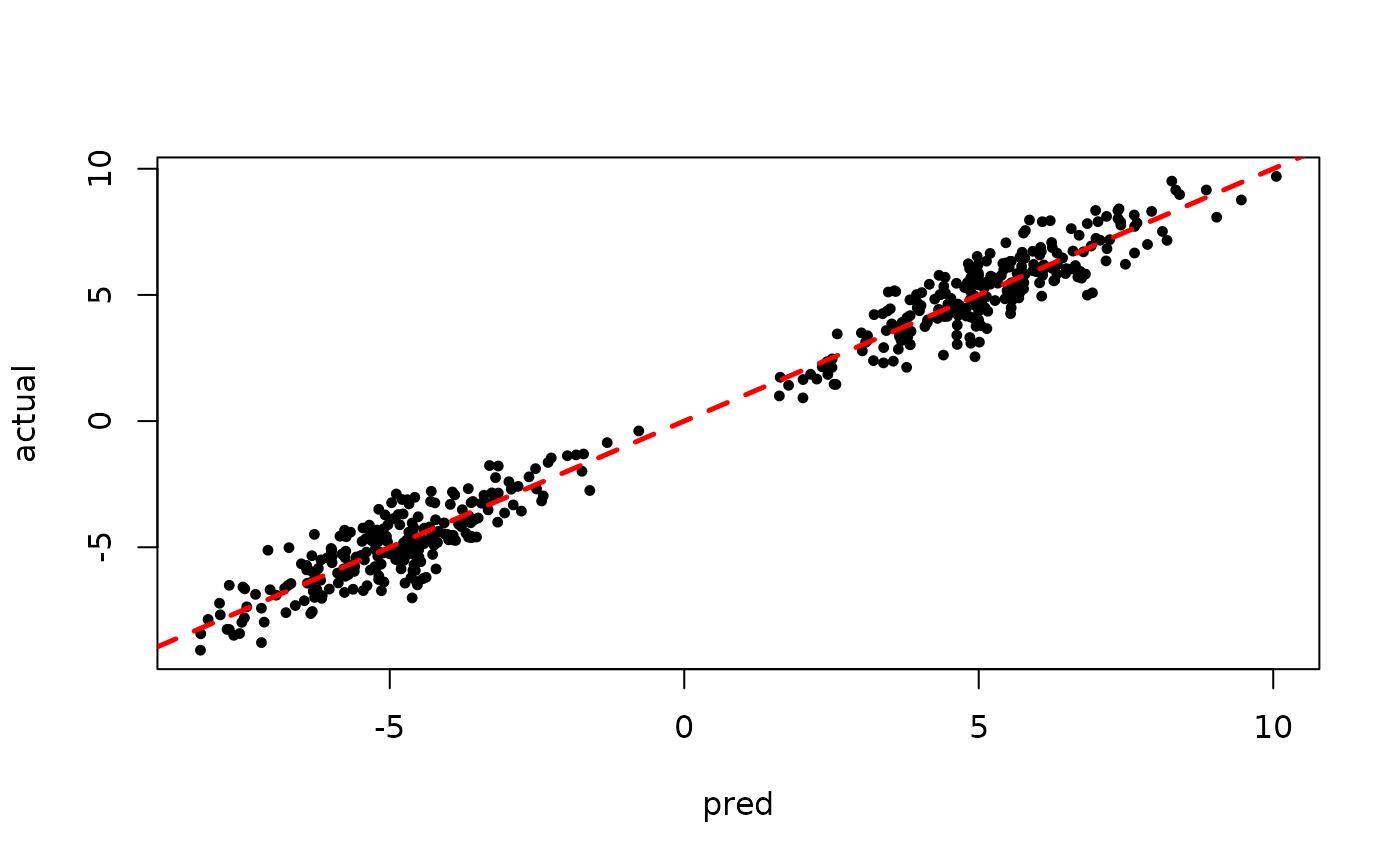
Inspect the BART samples obtained after “warm-starting” plus an additive random effects model
plot(sigma_samples[(num_warmstart+1):num_samples], ylab="sigma")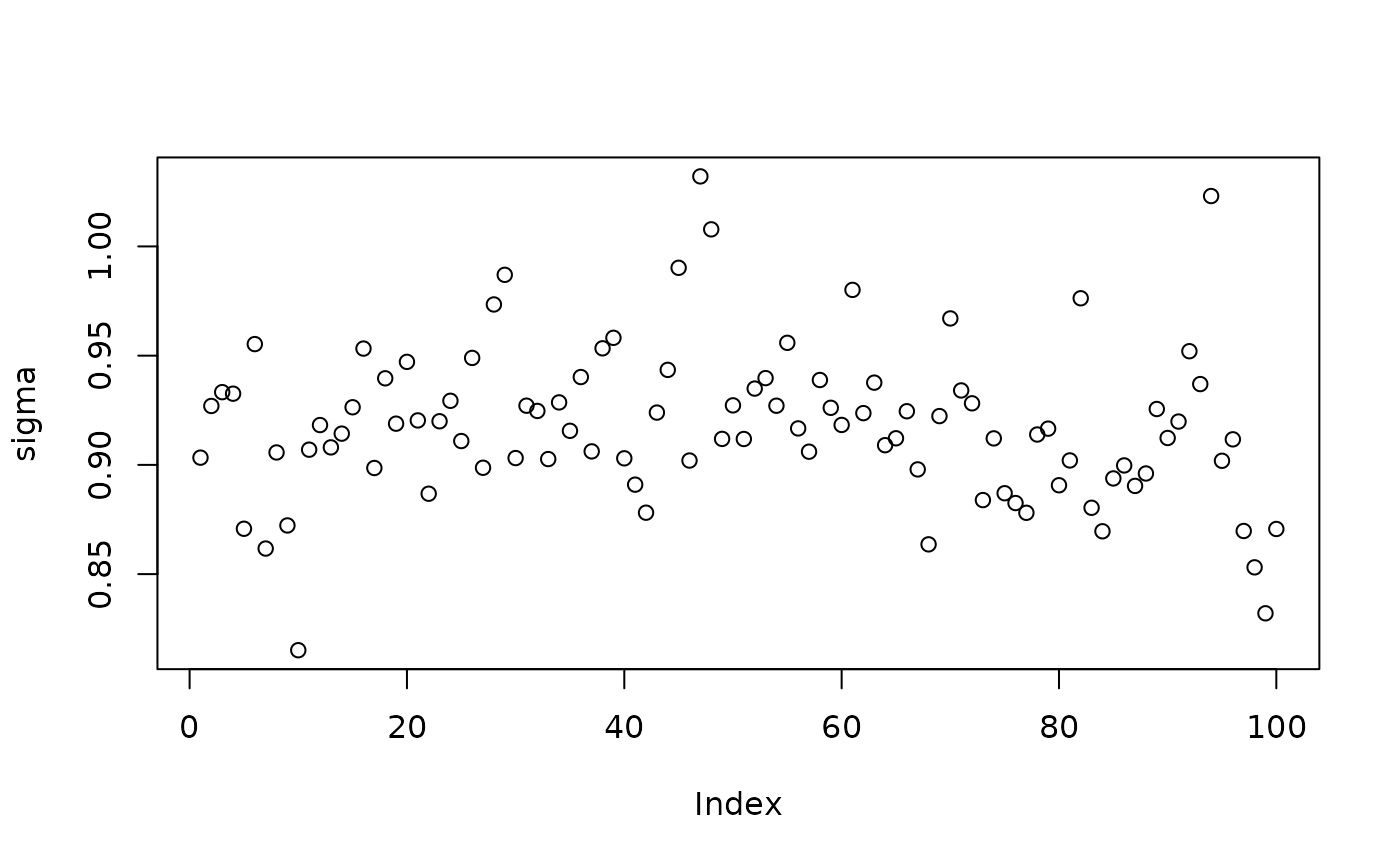
plot(rowMeans(preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)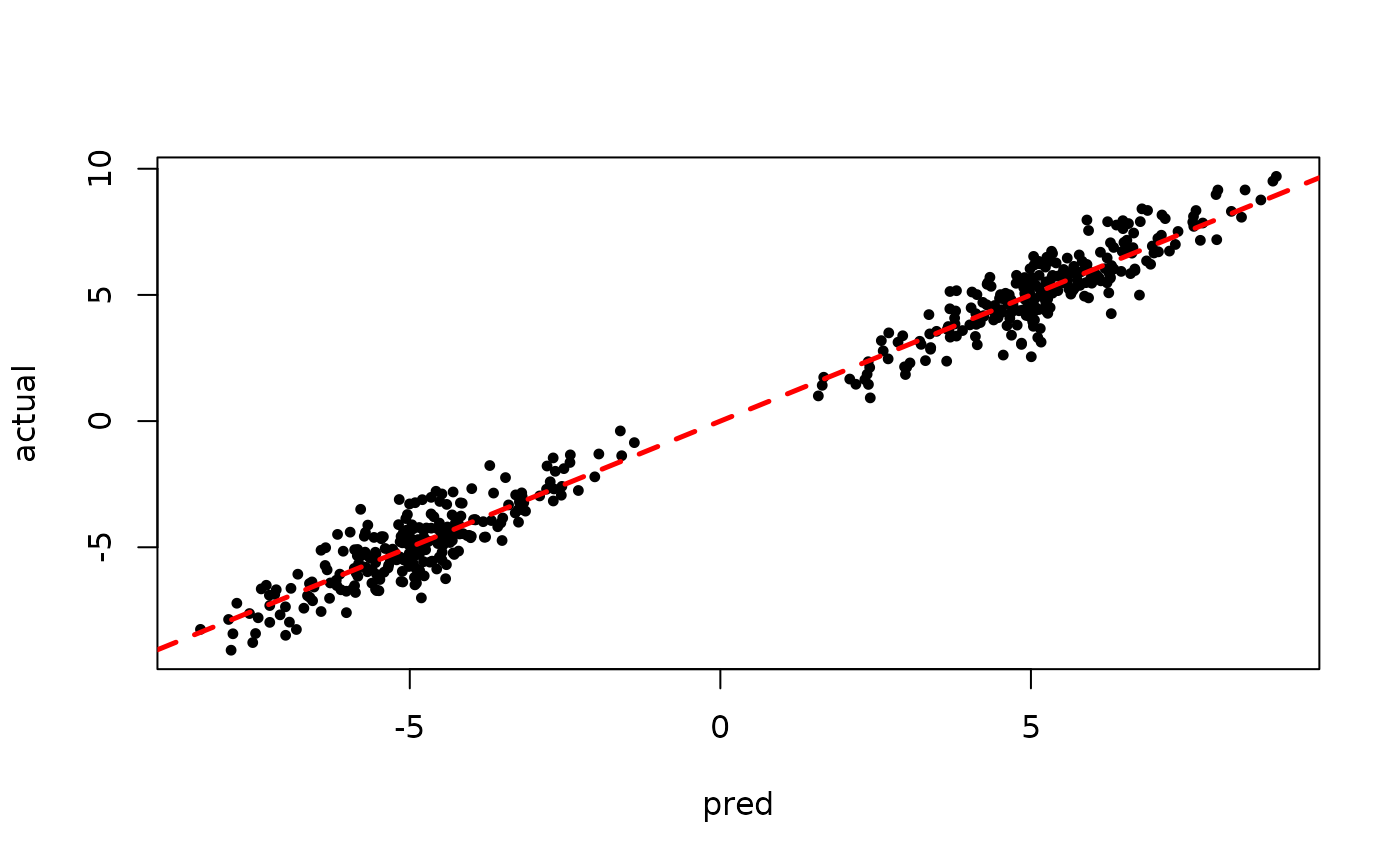
Now inspect the samples from the BART forest alone (without considering the random effect predictions)
plot(rowMeans(forest_preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)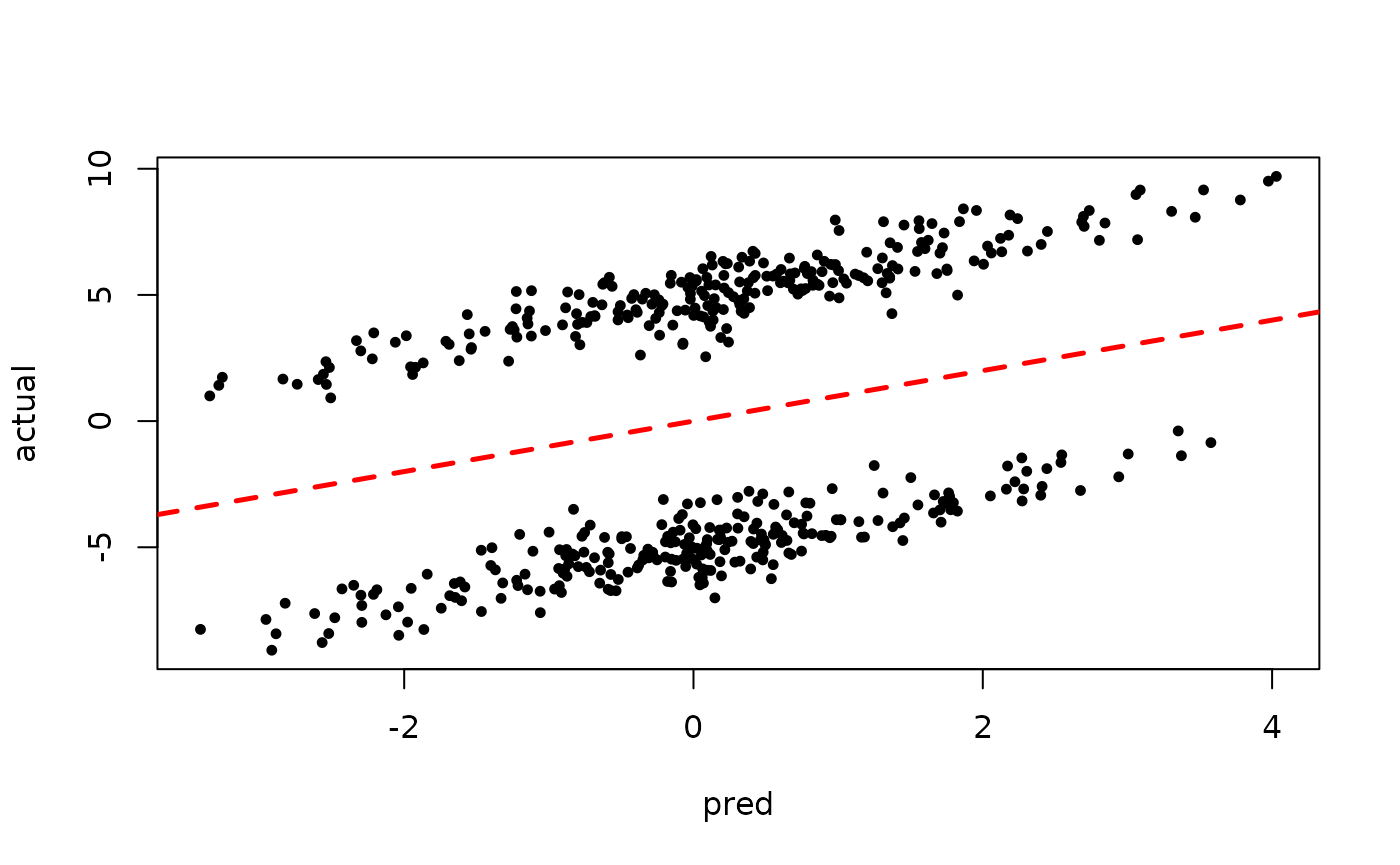
Demo 3: Supervised Learning with Additive Multi-Component Random Effects
We build once again on the above example, in this case allowing for a random intercept and regression coefficient (on a pre-specified basis) for each group (1 and 2).
Simulation
Simulate a partitioned linear model with a simple additive group random effect structure
# Generate the data
n <- 500
p_X <- 10
p_W <- 1
X <- matrix(runif(n*p_X), ncol = p_X)
W <- matrix(runif(n*p_W), ncol = p_W)
group_ids <- rep(c(1,2), n %/% 2)
rfx_coefs <- matrix(c(-5, -3, 5, 3), nrow=2, byrow=TRUE)
rfx_basis <- cbind(1, runif(n, -1, 1))
f_XW <- (
((0 <= X[,1]) & (0.25 > X[,1])) * (-3*W[,1]) +
((0.25 <= X[,1]) & (0.5 > X[,1])) * (-1*W[,1]) +
((0.5 <= X[,1]) & (0.75 > X[,1])) * (1*W[,1]) +
((0.75 <= X[,1]) & (1 > X[,1])) * (3*W[,1])
)
rfx_term <- rowSums(rfx_coefs[group_ids,] * rfx_basis)
y <- f_XW + rfx_term + rnorm(n, 0, 1)
# Standardize outcome
y_bar <- mean(y)
y_std <- sd(y)
resid <- (y-y_bar)/y_stdSampling
Set some parameters that inform the forest and variance parameter samplers
alpha <- 0.9
beta <- 1.25
min_samples_leaf <- 1
max_depth <- 10
num_trees <- 100
cutpoint_grid_size = 100
global_variance_init = 1.
tau_init = 0.5
leaf_prior_scale = matrix(c(tau_init), ncol = 1)
nu <- 4
lambda <- 0.5
a_leaf <- 2.
b_leaf <- 0.5
leaf_regression <- T
feature_types <- as.integer(rep(0, p_X)) # 0 = numeric
var_weights <- rep(1/p_X, p_X)Set some parameters that inform the random effects samplers
alpha_init <- c(1,0)
xi_init <- matrix(c(1,0,1,0),2,2)
sigma_alpha_init <- diag(1,2,2)
sigma_xi_init <- diag(1,2,2)
sigma_xi_shape <- 1
sigma_xi_scale <- 1Initialize R-level access to the C++ classes needed to sample our model
# Data
if (leaf_regression) {
forest_dataset <- createForestDataset(X, W)
outcome_model_type <- 1
} else {
forest_dataset <- createForestDataset(X)
outcome_model_type <- 0
}
outcome <- createOutcome(resid)
# Random number generator (std::mt19937)
rng <- createRNG()
# Sampling data structures
forest_model <- createForestModel(forest_dataset, feature_types,
num_trees, n, alpha, beta,
min_samples_leaf, max_depth)
# "Active forest" (which gets updated by the sample) and
# container of forest samples (which is written to when
# a sample is not discarded due to burn-in / thinning)
if (leaf_regression) {
forest_samples <- createForestContainer(num_trees, 1, F)
active_forest <- createForest(num_trees, 1, F)
} else {
forest_samples <- createForestContainer(num_trees, 1, T)
active_forest <- createForest(num_trees, 1, T)
}
# Random effects dataset
rfx_basis <- as.matrix(rfx_basis)
group_ids <- as.integer(group_ids)
rfx_dataset <- createRandomEffectsDataset(group_ids, rfx_basis)
# Random effects details
num_groups <- length(unique(group_ids))
num_components <- ncol(rfx_basis)
# Random effects tracker
rfx_tracker <- createRandomEffectsTracker(group_ids)
# Random effects model
rfx_model <- createRandomEffectsModel(num_components, num_groups)
rfx_model$set_working_parameter(alpha_init)
rfx_model$set_group_parameters(xi_init)
rfx_model$set_working_parameter_cov(sigma_alpha_init)
rfx_model$set_group_parameter_cov(sigma_xi_init)
rfx_model$set_variance_prior_shape(sigma_xi_shape)
rfx_model$set_variance_prior_scale(sigma_xi_scale)
# Random effect samples
rfx_samples <- createRandomEffectSamples(num_components, num_groups, rfx_tracker)Prepare to run the sampler
num_warmstart <- 10
num_mcmc <- 100
num_samples <- num_warmstart + num_mcmc
global_var_samples <- c(global_variance_init, rep(0, num_samples))
leaf_scale_samples <- c(tau_init, rep(0, num_samples))Run the grow-from-root sampler to “warm-start” BART
for (i in 1:num_warmstart) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = T
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
# Sample random effects model
rfx_model$sample_random_effect(rfx_dataset, outcome, rfx_tracker, rfx_samples,
TRUE, global_var_samples[i+1], rng)
}Pick up from the last GFR forest (and associated global variance / leaf scale parameters) with an MCMC sampler
for (i in (num_warmstart+1):num_samples) {
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = F
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
# Sample leaf node variance parameter and update `leaf_prior_scale`
leaf_scale_samples[i+1] <- sample_tau_one_iteration(
active_forest, rng, a_leaf, b_leaf
)
leaf_prior_scale[1,1] <- leaf_scale_samples[i+1]
# Sample random effects model
rfx_model$sample_random_effect(rfx_dataset, outcome, rfx_tracker, rfx_samples,
TRUE, global_var_samples[i+1], rng)
}Predict and rescale samples
# Forest predictions
forest_preds <- forest_samples$predict(forest_dataset)*y_std + y_bar
# Random effects predictions
rfx_preds <- rfx_samples$predict(group_ids, rfx_basis)*y_std
# Overall predictions
preds <- forest_preds + rfx_preds
# Global error variance
sigma_samples <- sqrt(global_var_samples)*y_stdResults
Inspect the initial samples obtained via grow-from-root and an additive random effects model
plot(sigma_samples[1:num_warmstart], ylab="sigma")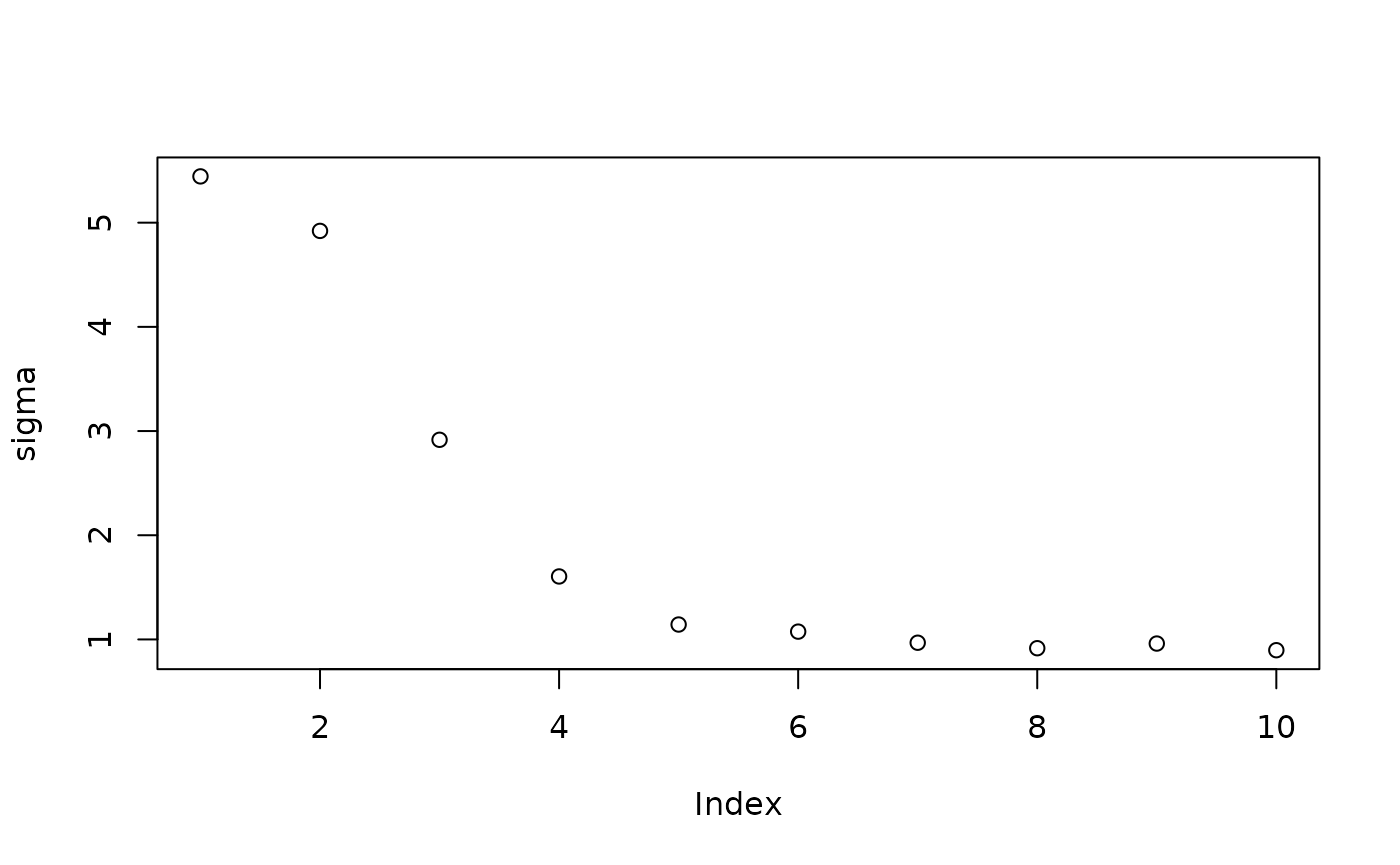
plot(rowMeans(preds[,1:num_warmstart]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)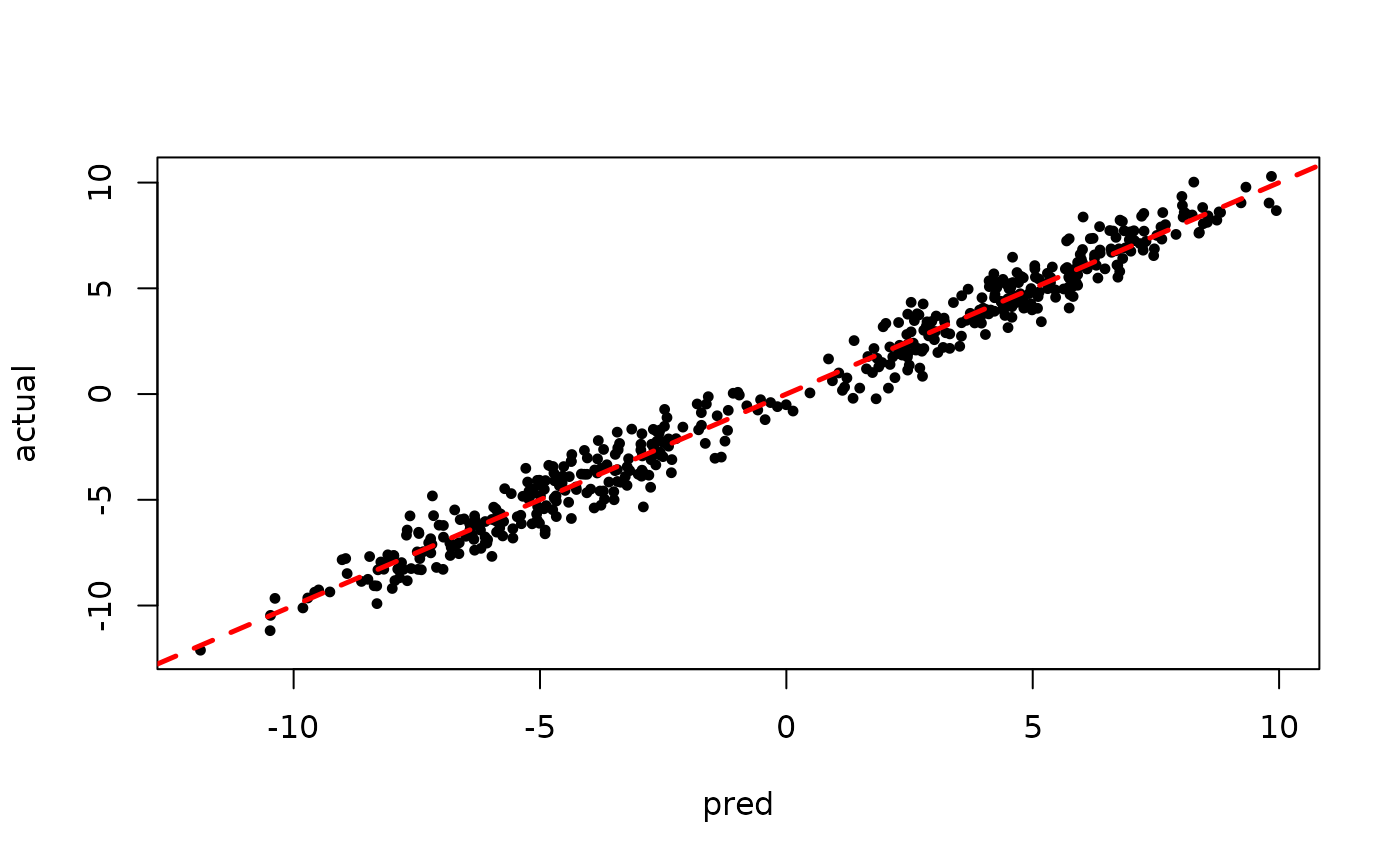
Inspect the BART samples obtained after “warm-starting” plus an additive random effects model
plot(sigma_samples[(num_warmstart+1):num_samples], ylab="sigma")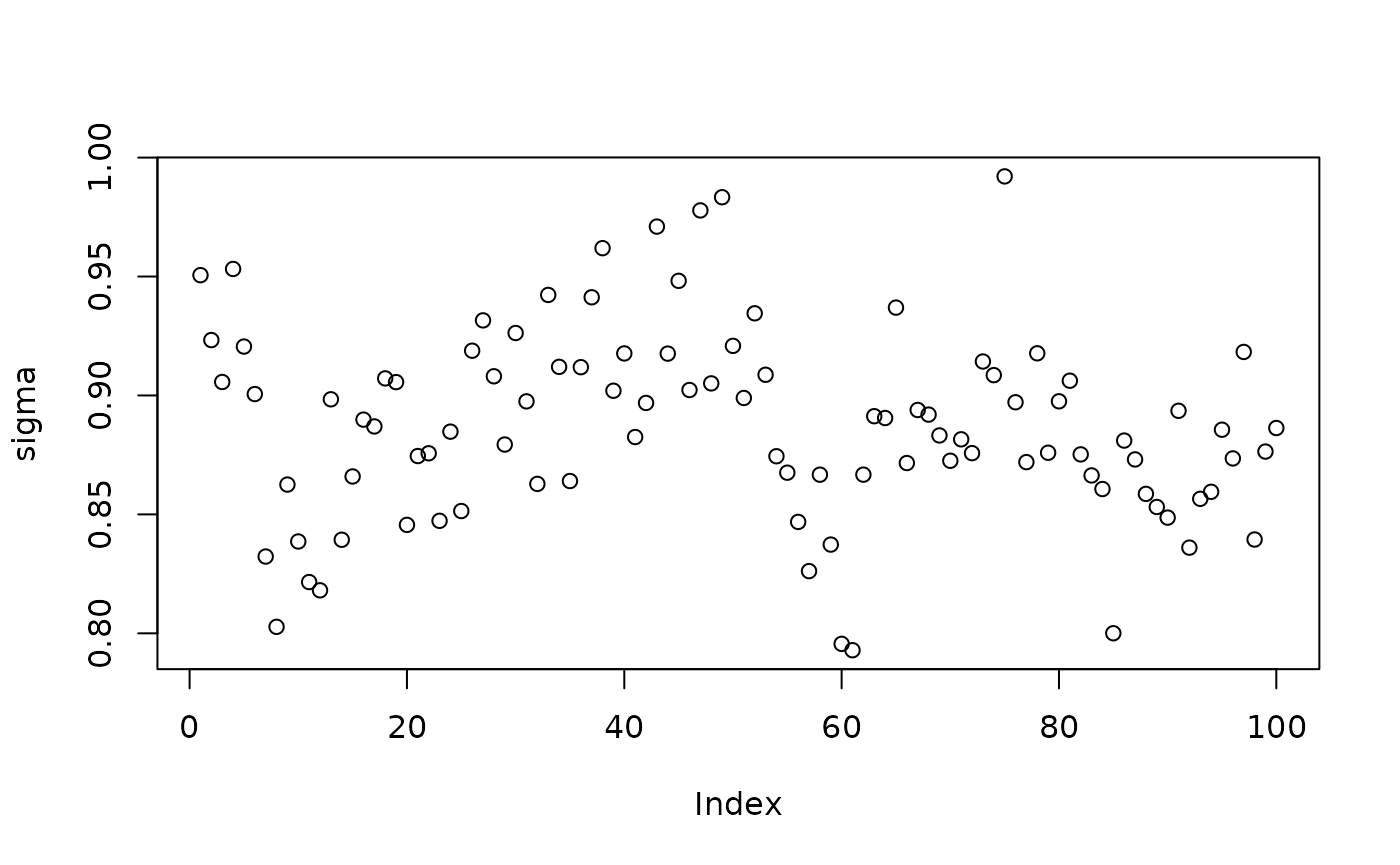
plot(rowMeans(preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)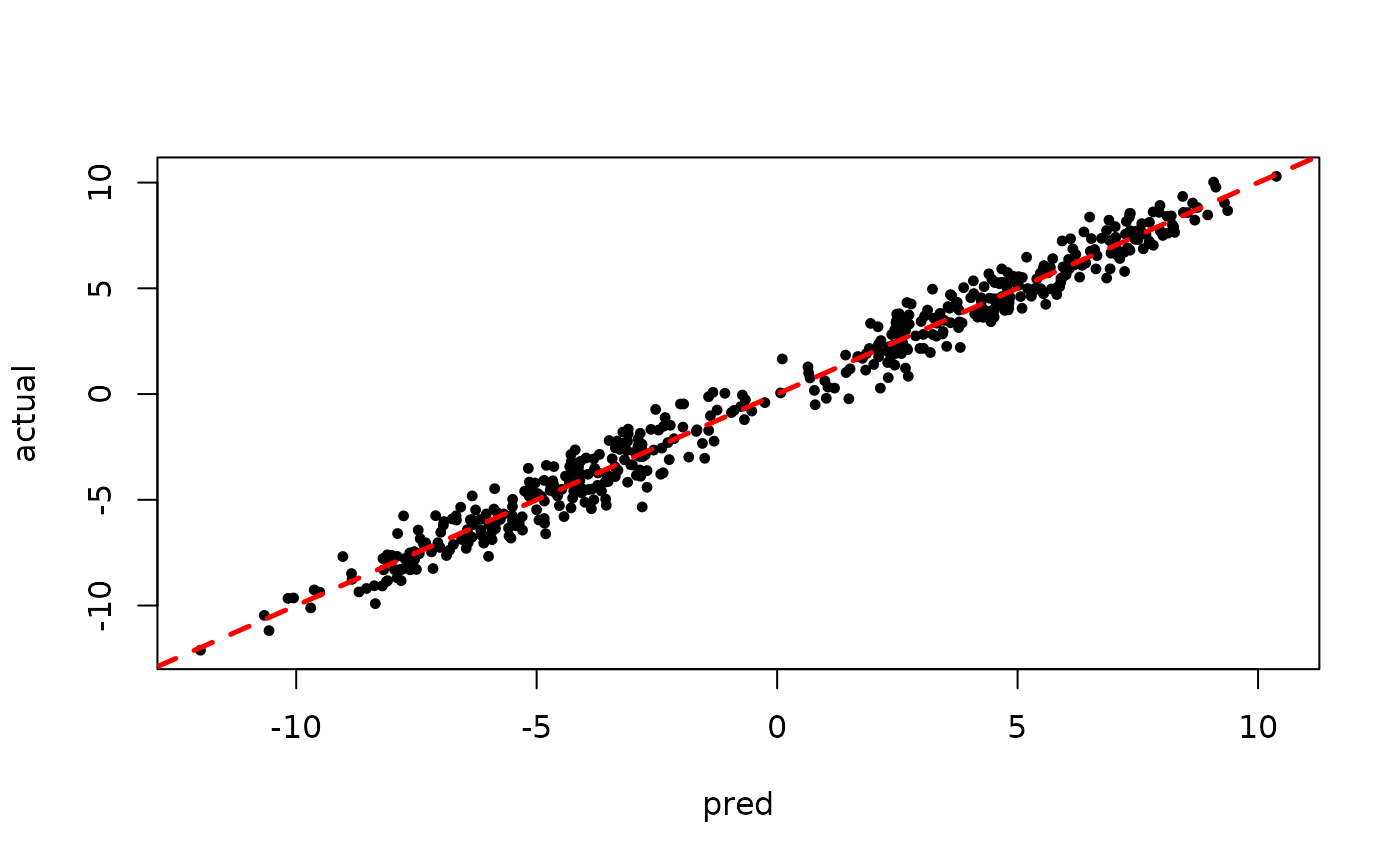
Now inspect the samples from the BART forest alone (without considering the random effect predictions)
plot(rowMeans(forest_preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)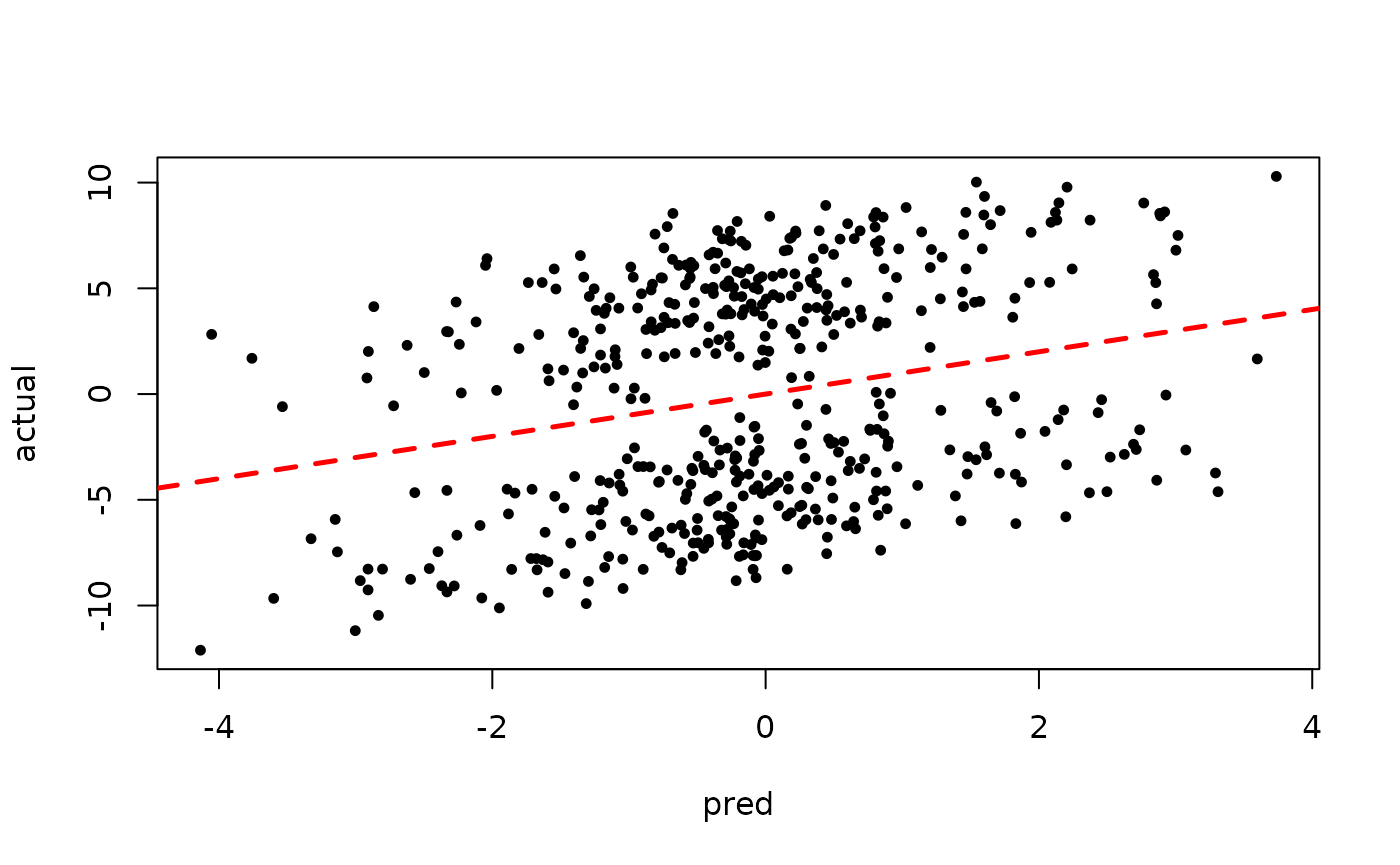
Demo 4: Supervised Learning with Additive Linear Model
Instead of group random effects, here we show how to combine a stochastic forest with an additive linear regression term. The model corresponds broadly to
Simulation
Simulate a partitioned linear model with a simple additive group random effect structure
# Generate the data
n <- 500
p_X <- 10
p_W <- 1
X <- matrix(runif(n*p_X), ncol = p_X)
W <- matrix(runif(n*p_W), ncol = p_W)
beta_W <- c(5)
f_XW <- (
((0 <= X[,1]) & (0.25 > X[,1])) * (-3) +
((0.25 <= X[,1]) & (0.5 > X[,1])) * (-1) +
((0.5 <= X[,1]) & (0.75 > X[,1])) * (1) +
((0.75 <= X[,1]) & (1 > X[,1])) * (3)
)
lm_term <- W %*% beta_W
y <- lm_term + f_XW + rnorm(n, 0, 1)
# Standardize outcome
y_bar <- mean(y)
y_std <- sd(y)
resid <- (y-y_bar)/y_stdSampling
Set some parameters that inform the forest and variance parameter samplers
alpha_bart <- 0.9
beta_bart <- 1.25
min_samples_leaf <- 1
max_depth <- 10
num_trees <- 100
cutpoint_grid_size = 100
global_variance_init = 1.
tau_init = 0.5
leaf_prior_scale = matrix(c(tau_init), ncol = 1)
nu <- 4
lambda <- 0.5
a_leaf <- 2.
b_leaf <- 0.5
leaf_regression <- F
feature_types <- as.integer(rep(0, p_X)) # 0 = numeric
var_weights <- rep(1/p_X, p_X)
beta_tau <- 20Initialize R-level access to the C++ classes needed to sample our model
# Data
if (leaf_regression) {
forest_dataset <- createForestDataset(X, W)
outcome_model_type <- 1
} else {
forest_dataset <- createForestDataset(X)
outcome_model_type <- 0
}
outcome <- createOutcome(resid)
# Random number generator (std::mt19937)
rng <- createRNG()
# Sampling data structures
forest_model <- createForestModel(forest_dataset, feature_types,
num_trees, n, alpha_bart, beta_bart,
min_samples_leaf, max_depth)
# "Active forest" (which gets updated by the sample) and
# container of forest samples (which is written to when
# a sample is not discarded due to burn-in / thinning)
if (leaf_regression) {
forest_samples <- createForestContainer(num_trees, 1, F)
active_forest <- createForest(num_trees, 1, F)
} else {
forest_samples <- createForestContainer(num_trees, 1, T)
active_forest <- createForest(num_trees, 1, T)
}Prepare to run the sampler
num_warmstart <- 20
num_mcmc <- 100
num_samples <- num_warmstart + num_mcmc
beta_init <- 0
global_var_samples <- c(global_variance_init, rep(0, num_samples))
leaf_scale_samples <- c(tau_init, rep(0, num_samples))
beta_samples <- c(beta_init, rep(0, num_samples))Run the grow-from-root sampler to “warm-start” BART
for (i in 1:num_warmstart) {
# Initialize vectors needed for posterior sampling
if (i == 1) {
beta_hat <- beta_init
yhat_forest <- rep(0, n)
partial_res <- resid - yhat_forest
} else {
yhat_forest <- forest_samples$predict_raw_single_forest(forest_dataset, (i-1)-1)
partial_res <- resid - yhat_forest
outcome$add_vector(W %*% beta_hat)
}
# Sample beta from bayesian linear model with gaussian prior
sigma2 <- global_var_samples[i]
beta_posterior_mean <- sum(partial_res*W[,1])/(sigma2 + sum(W[,1]*W[,1]))
beta_posterior_var <- (sigma2*beta_tau)/(sigma2 + sum(W[,1]*W[,1]))
beta_hat <- rnorm(1, beta_posterior_mean, sqrt(beta_posterior_var))
beta_samples[i+1] <- beta_hat
# Update partial residual before sampling forest
outcome$subtract_vector(W %*% beta_hat)
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, sigma2, cutpoint_grid_size, keep_forest = T, gfr = T
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
}Pick up from the last GFR forest (and associated global variance / leaf scale parameters) with an MCMC sampler
for (i in (num_warmstart+1):num_samples) {
# Initialize vectors needed for posterior sampling
if (i == 1) {
beta_hat <- beta_init
yhat_forest <- rep(0, n)
partial_res <- resid - yhat_forest
} else {
yhat_forest <- forest_samples$predict_raw_single_forest(forest_dataset, (i-1)-1)
partial_res <- resid - yhat_forest
outcome$add_vector(W %*% beta_hat)
}
# Sample beta from bayesian linear model with gaussian prior
sigma2 <- global_var_samples[i]
beta_posterior_mean <- sum(partial_res*W[,1])/(sigma2 + sum(W[,1]*W[,1]))
beta_posterior_var <- (sigma2*beta_tau)/(sigma2 + sum(W[,1]*W[,1]))
beta_hat <- rnorm(1, beta_posterior_mean, sqrt(beta_posterior_var))
beta_samples[i+1] <- beta_hat
# Update partial residual before sampling forest
outcome$subtract_vector(W %*% beta_hat)
# Sample forest
forest_model$sample_one_iteration(
forest_dataset, outcome, forest_samples, active_forest, rng, feature_types,
outcome_model_type, leaf_prior_scale, var_weights,
1, 1, global_var_samples[i], cutpoint_grid_size, keep_forest = T, gfr = F
)
# Sample global variance parameter
global_var_samples[i+1] <- sample_sigma2_one_iteration(
outcome, forest_dataset, rng, nu, lambda
)
}Predict and rescale samples
# Linear model predictions
lm_preds <- (sapply(1:num_samples, function(x) W[,1]*beta_samples[x+1]))*y_std
# Forest predictions
forest_preds <- forest_samples$predict(forest_dataset)*y_std + y_bar
# Overall predictions
preds <- forest_preds + lm_preds
# Global error variance
sigma_samples <- sqrt(global_var_samples)*y_std
# Regression parameter
beta_samples <- beta_samples*y_stdResults
Inspect the initial samples obtained via grow-from-root and an additive random effects model
plot(sigma_samples[1:num_warmstart], ylab="sigma")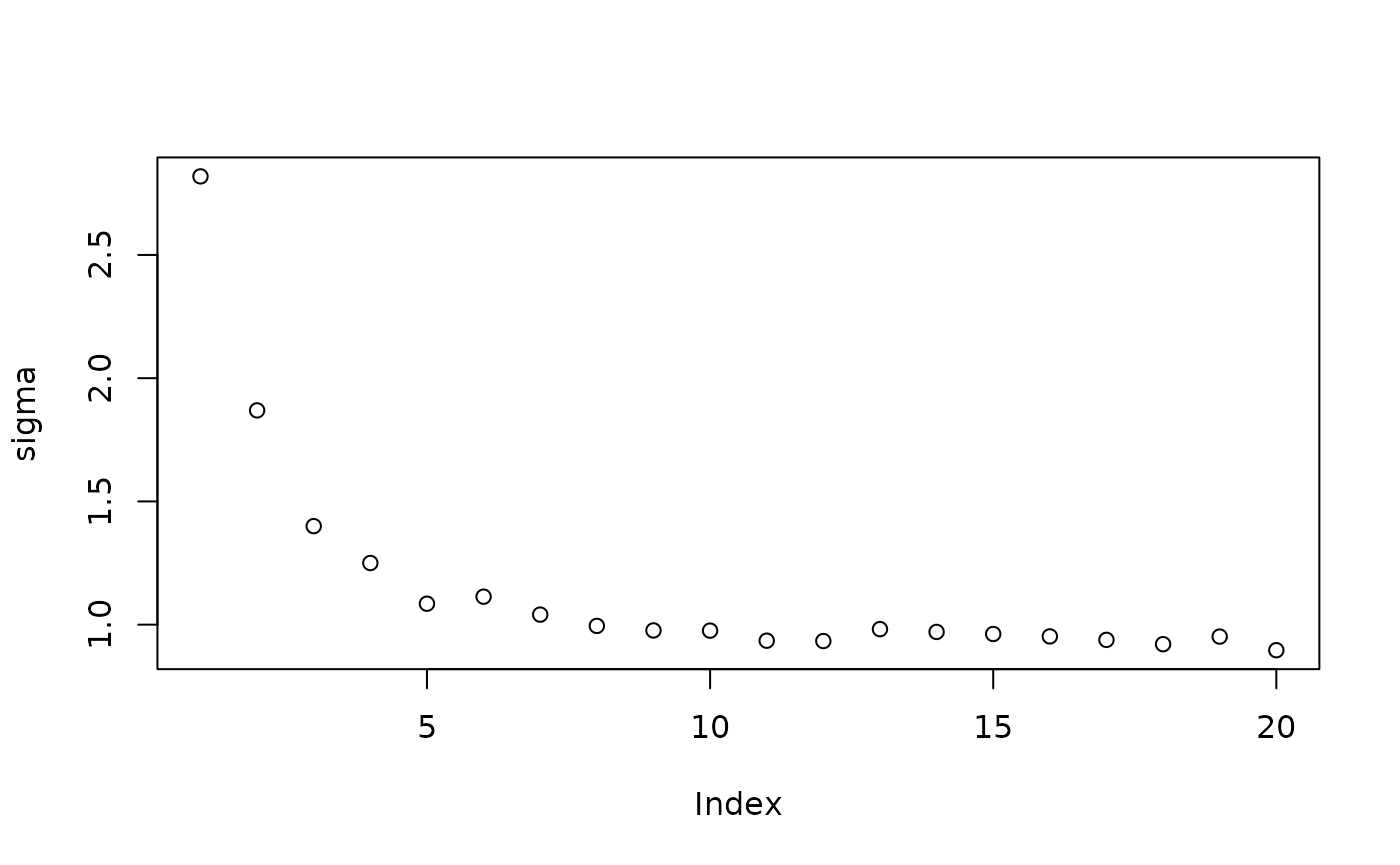
plot(beta_samples[1:num_warmstart], ylab="beta")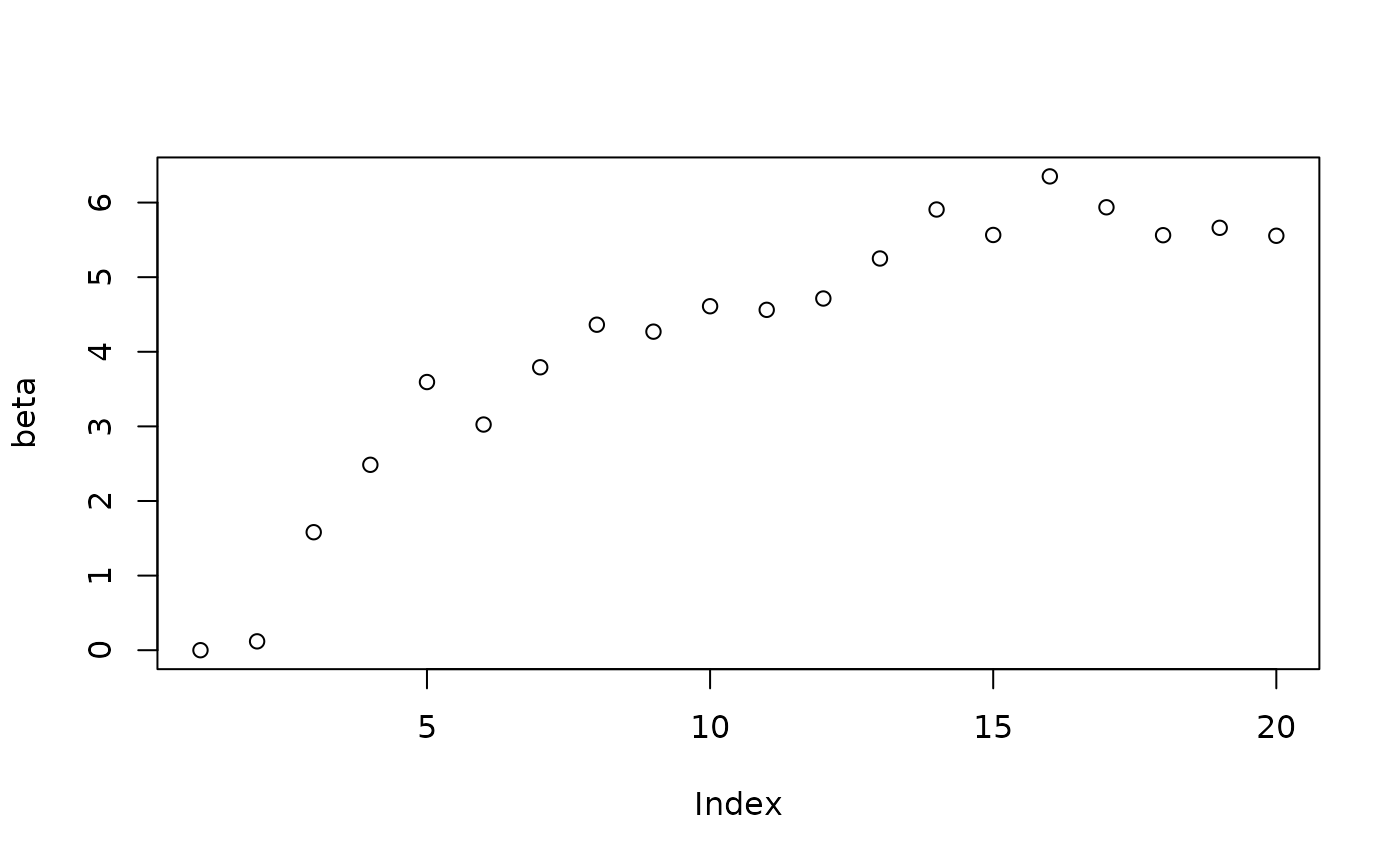
plot(rowMeans(preds[,1:num_warmstart]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)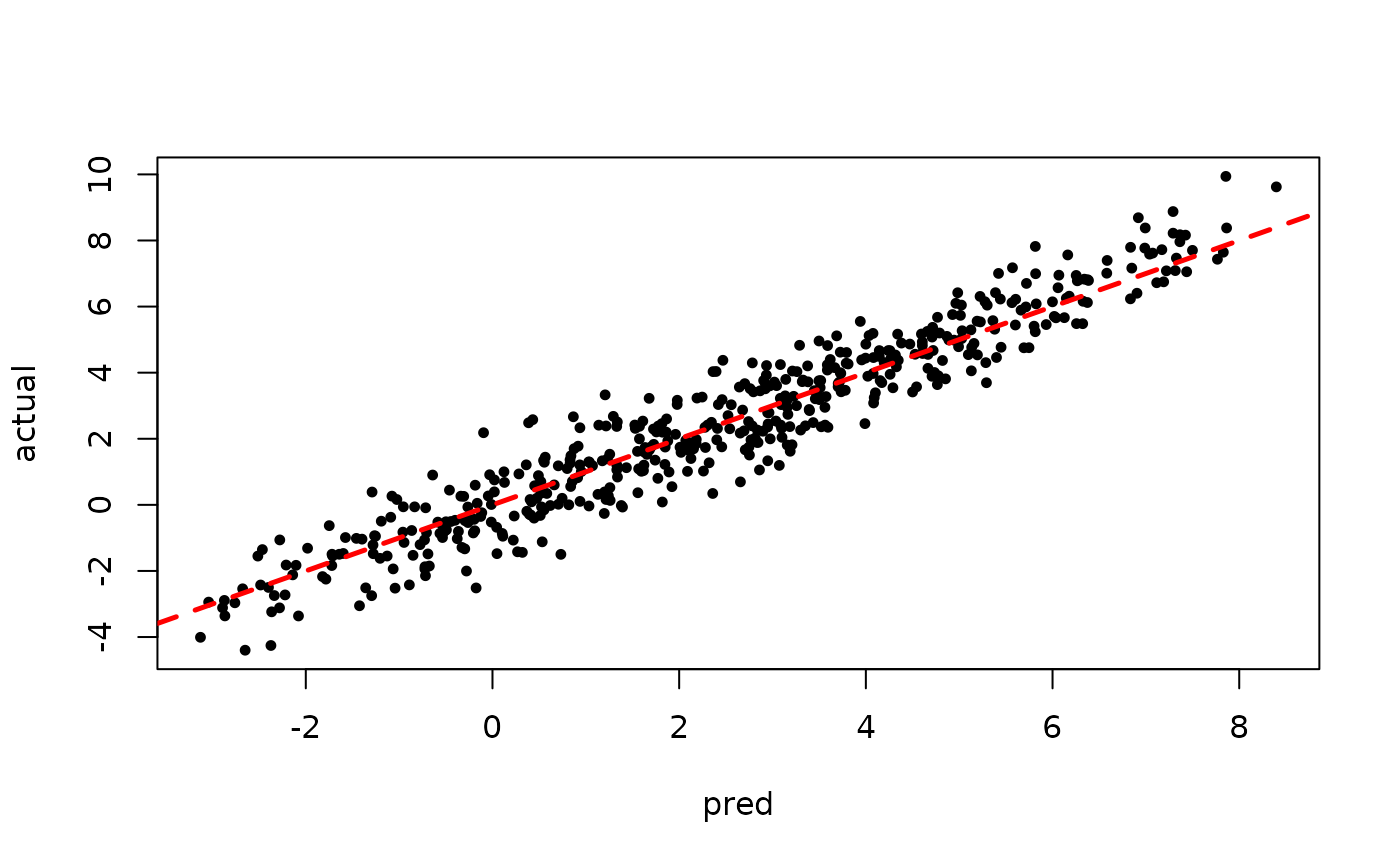
Inspect the BART samples obtained after “warm-starting” plus an additive random effects model
plot(sigma_samples[(num_warmstart+1):num_samples], ylab="sigma")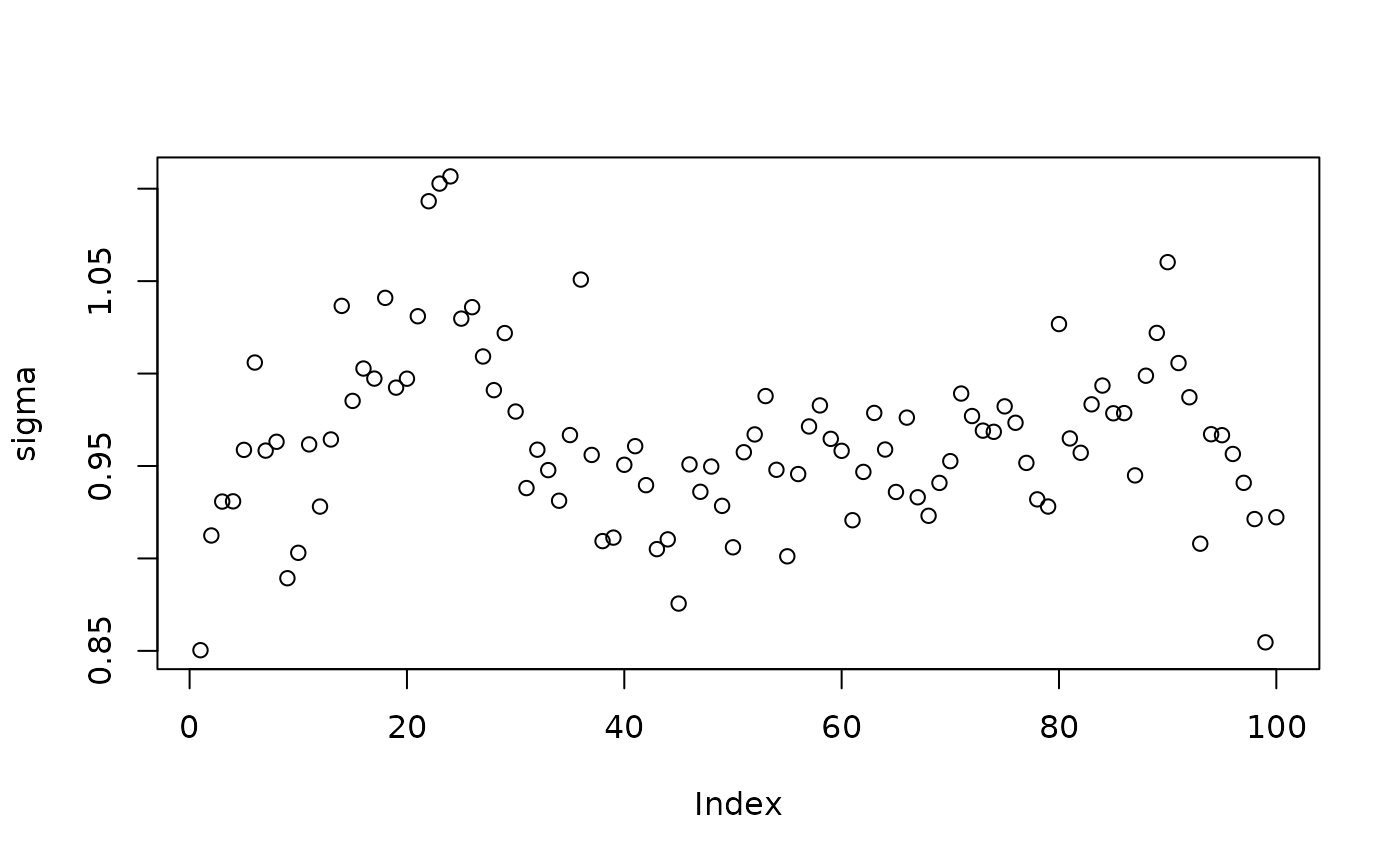
plot(beta_samples[(num_warmstart+1):num_samples], ylab="beta")
plot(rowMeans(preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)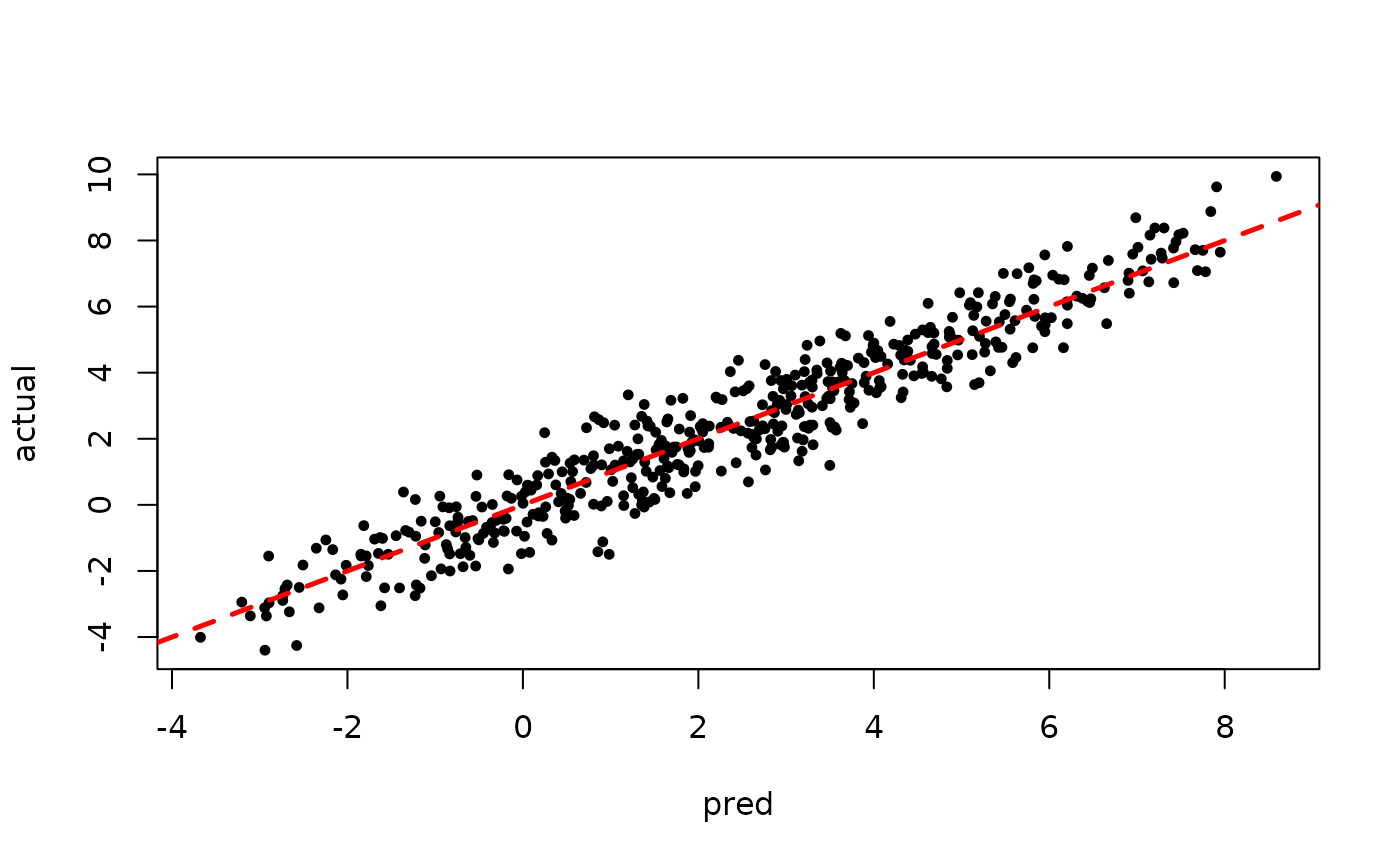
Now inspect the samples from the BART forest alone (without considering the additive linear model predictions)
plot(rowMeans(forest_preds[,(num_warmstart+1):num_samples]), y, pch=16,
cex=0.75, xlab = "pred", ylab = "actual")
abline(0,1,col="red",lty=2,lwd=2.5)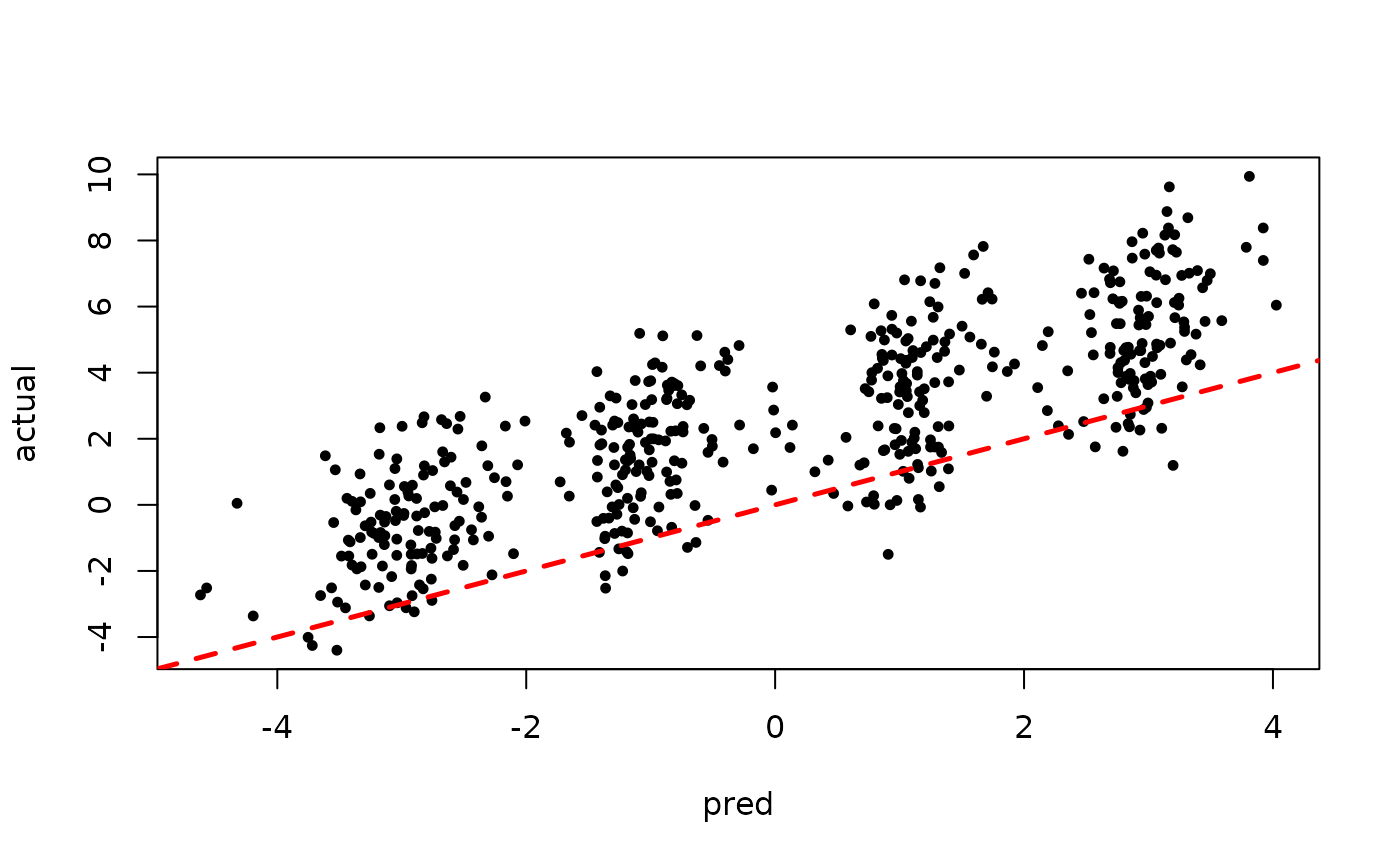
Demo 5: Causal Inference
Here we show how to implement the Bayesian Causal Forest (BCF) model
of Hahn, Murray, and Carvalho (2020) using
stochtree’s prototype API, including demoing a non-trivial
sampling step done at the R level.
Background
While the supervised learning case of the previous demo is conceptually simple, we motivate the causal effect estimation task with some additional notation. Let refer to a continuous outcome of interest, refer to a binary treatment, and to a set of covariates that may influence , , or both.
If is an exhaustive set of covariates that influence and , we can specific in terms of a causal model (see for example Pearl (2009)) as where is outcome specific random noise and is a function that generates (in many cases, can be thought of as the inverse of the CDF conditional on and ).
The “potential outcomes” (see Imbens and Rubin (2015)) can be recovered by and .
The causal outcome model can be decomposed into “mean” and “error” terms as below
Here is precisely the conditional average treatment effect (CATE) estimand. Unfortunately, the functional form of is unavailable for analysis, so that cannot be derived.
This is where flexible, regularized nonparametrics enter the picture, as we aim to estimate and from data.
Bayesian Causal Forest (BCF)
BCF estimates and using separate BART forests for each term. Furthermore, rather than rely on the common implicit coding of as 0 for control observations and 1 for treated observations, they consider coding control observations with a parameter and treated observations with a parameter . Placing a prior on each , this essentially redefines the outcome model as
Updating each requires an additional Gibbs step, which we derive here. Conditioning on sampled forests and , we are essentially regressing on which has a closed form posterior where and .
Simulation
The simulated causal DGP mirrors the nonlinear, heterogeneous treatment effect DGP presented in Hahn, Murray, and Carvalho (2020).
n <- 500
x1 <- rnorm(n)
x2 <- rnorm(n)
x3 <- rnorm(n)
x4 <- as.numeric(rbinom(n,1,0.5))
x5 <- as.numeric(sample(1:3,n,replace=TRUE))
X <- cbind(x1,x2,x3,x4,x5)
p <- ncol(X)
g <- function(x) {ifelse(x[,5]==1,2,ifelse(x[,5]==2,-1,4))}
mu1 <- function(x) {1+g(x)+x[,1]*x[,3]}
mu2 <- function(x) {1+g(x)+6*abs(x[,3]-1)}
tau1 <- function(x) {rep(3,nrow(x))}
tau2 <- function(x) {1+2*x[,2]*x[,4]}
mu_x <- mu1(X)
tau_x <- tau2(X)
pi_x <- 0.8*pnorm((3*mu_x/sd(mu_x)) - 0.5*X[,1]) + 0.05 + runif(n)/10
Z <- rbinom(n,1,pi_x)
E_XZ <- mu_x + Z*tau_x
snr <- 4
y <- E_XZ + rnorm(n, 0, 1)*(sd(E_XZ)/snr)
# Standardize outcome
y_bar <- mean(y)
y_std <- sd(y)
resid <- (y-y_bar)/y_stdSampling
Set some parameters that inform the forest and variance parameter samplers
# Mu forest
alpha_mu <- 0.95
beta_mu <- 2.0
min_samples_leaf_mu <- 5
max_depth_mu <- 10
num_trees_mu <- 250
cutpoint_grid_size_mu = 100
tau_init_mu = 1/num_trees_mu
leaf_prior_scale_mu = matrix(c(tau_init_mu), ncol = 1)
a_leaf_mu <- 3.
b_leaf_mu <- var(resid)/(num_trees_mu)
leaf_regression_mu <- F
sigma_leaf_mu <- var(resid)/(num_trees_mu)
current_leaf_scale_mu <- as.matrix(sigma_leaf_mu)
# Tau forest
alpha_tau <- 0.25
beta_tau <- 3.0
min_samples_leaf_tau <- 5
max_depth_tau <- 10
num_trees_tau <- 50
cutpoint_grid_size_tau = 100
a_leaf_tau <- 3.
b_leaf_tau <- var(resid)/(2*num_trees_tau)
leaf_regression_tau <- T
sigma_leaf_tau <- var(resid)/(2*num_trees_tau)
current_leaf_scale_tau <- as.matrix(sigma_leaf_tau)
# Common parameters
nu <- 3
sigma2hat <- (sigma(lm(resid~X)))^2
quantile_cutoff <- 0.9
if (is.null(lambda)) {
lambda <- (sigma2hat*qgamma(1-quantile_cutoff,nu))/nu
}
sigma2 <- sigma2hat
current_sigma2 <- sigma2Prepare to run the sampler (now we must specify initial values for and , for which we choose -1/2 and 1/2 instead of 0 and 1).
# Sampling composition
num_gfr <- 20
num_burnin <- 0
num_mcmc <- 100
num_samples <- num_gfr + num_burnin + num_mcmc
# Sigma^2 samples
global_var_samples <- rep(0, num_samples)
# Adaptive coding parameter samples
b_0_samples <- rep(0, num_samples)
b_1_samples <- rep(0, num_samples)
b_0 <- -0.5
b_1 <- 0.5
current_b_0 <- b_0
current_b_1 <- b_1
tau_basis <- (1-Z)*current_b_0 + Z*current_b_1Initialize R-level access to the C++ classes needed to sample our model
# Data
X_mu <- cbind(X, pi_x)
X_tau <- X
feature_types <- c(0,0,0,1,1)
feature_types_mu <- as.integer(c(feature_types,0))
feature_types_tau <- as.integer(feature_types)
variable_weights_mu = rep(1/ncol(X_mu), ncol(X_mu))
variable_weights_tau = rep(1/ncol(X_tau), ncol(X_tau))
forest_dataset_mu <- createForestDataset(X_mu)
forest_dataset_tau <- createForestDataset(X_tau, tau_basis)
outcome <- createOutcome(resid)
# Random number generator (std::mt19937)
rng <- createRNG()
# Sampling data structures
forest_model_mu <- createForestModel(
forest_dataset_mu, feature_types_mu, num_trees_mu, nrow(X_mu),
alpha_mu, beta_mu, min_samples_leaf_mu, max_depth_mu
)
forest_model_tau <- createForestModel(
forest_dataset_tau, feature_types_tau, num_trees_tau, nrow(X_tau),
alpha_tau, beta_tau, min_samples_leaf_tau, max_depth_tau
)
# Container of forest samples
forest_samples_mu <- createForestContainer(num_trees_mu, 1, T)
active_forest_mu <- createForest(num_trees_mu, 1, T)
forest_samples_tau <- createForestContainer(num_trees_tau, 1, F)
active_forest_tau <- createForest(num_trees_tau, 1, F)
# Initialize the leaves of each tree in the prognostic forest
active_forest_mu$prepare_for_sampler(forest_dataset_mu, outcome, forest_model_mu, 0, mean(resid))
active_forest_mu$adjust_residual(forest_dataset_mu, outcome, forest_model_mu, F, F)
# Initialize the leaves of each tree in the treatment effect forest
active_forest_tau$prepare_for_sampler(forest_dataset_tau, outcome, forest_model_tau, 1, 0.)
active_forest_tau$adjust_residual(forest_dataset_tau, outcome, forest_model_tau, T, F)Run the grow-from-root sampler to “warm-start” BART, also updating the adaptive coding parameter and
if (num_gfr > 0){
for (i in 1:num_gfr) {
# Sample the prognostic forest
forest_model_mu$sample_one_iteration(
forest_dataset_mu, outcome, forest_samples_mu, active_forest_mu, rng,
feature_types_mu, 0, current_leaf_scale_mu, variable_weights_mu,
1, 1, current_sigma2, cutpoint_grid_size, keep_forest = T, gfr = T,
pre_initialized = T
)
# Sample variance parameters (if requested)
global_var_samples[i] <- sample_sigma2_one_iteration(
outcome, forest_dataset_mu, rng, nu, lambda
)
current_sigma2 <- global_var_samples[i]
# Sample the treatment forest
forest_model_tau$sample_one_iteration(
forest_dataset_tau, outcome, forest_samples_tau, active_forest_tau, rng,
feature_types_tau, 1, current_leaf_scale_tau, variable_weights_tau,
1, 1, current_sigma2, cutpoint_grid_size, keep_forest = T, gfr = T,
pre_initialized = T
)
# Sample adaptive coding parameters
mu_x_raw <- active_forest_mu$predict_raw(forest_dataset_mu)
tau_x_raw <- active_forest_tau$predict_raw(forest_dataset_tau)
s_tt0 <- sum(tau_x_raw*tau_x_raw*(Z==0))
s_tt1 <- sum(tau_x_raw*tau_x_raw*(Z==1))
partial_resid_mu <- resid - mu_x_raw
s_ty0 <- sum(tau_x_raw*partial_resid_mu*(Z==0))
s_ty1 <- sum(tau_x_raw*partial_resid_mu*(Z==1))
current_b_0 <- rnorm(1, (s_ty0/(s_tt0 + 2*current_sigma2)),
sqrt(current_sigma2/(s_tt0 + 2*current_sigma2)))
current_b_1 <- rnorm(1, (s_ty1/(s_tt1 + 2*current_sigma2)),
sqrt(current_sigma2/(s_tt1 + 2*current_sigma2)))
tau_basis <- (1-Z)*current_b_0 + Z*current_b_1
forest_dataset_tau$update_basis(tau_basis)
forest_model_tau$propagate_basis_update(forest_dataset_tau, outcome, active_forest_tau)
b_0_samples[i] <- current_b_0
b_1_samples[i] <- current_b_1
# Sample variance parameters (if requested)
global_var_samples[i] <- sample_sigma2_one_iteration(outcome, forest_dataset_tau, rng, nu, lambda)
current_sigma2 <- global_var_samples[i]
}
}Pick up from the last GFR forest (and associated global variance / leaf scale parameters) with an MCMC sampler
if (num_burnin + num_mcmc > 0) {
for (i in (num_gfr+1):num_samples) {
# Sample the prognostic forest
forest_model_mu$sample_one_iteration(
forest_dataset_mu, outcome, forest_samples_mu, active_forest_mu, rng, feature_types_mu,
0, current_leaf_scale_mu, variable_weights_mu, 1, 1, current_sigma2,
cutpoint_grid_size, keep_forest = T, gfr = F, pre_initialized = T
)
# Sample global variance parameter
global_var_samples[i] <- sample_sigma2_one_iteration(outcome, forest_dataset_mu, rng, nu, lambda)
current_sigma2 <- global_var_samples[i]
# Sample the treatment forest
forest_model_tau$sample_one_iteration(
forest_dataset_tau, outcome, forest_samples_tau, active_forest_tau, rng, feature_types_tau,
1, current_leaf_scale_tau, variable_weights_tau, 1, 1, current_sigma2,
cutpoint_grid_size, keep_forest = T, gfr = F, pre_initialized = T
)
# Sample coding parameters
mu_x_raw <- active_forest_mu$predict_raw(forest_dataset_mu)
tau_x_raw <- active_forest_tau$predict_raw(forest_dataset_tau)
s_tt0 <- sum(tau_x_raw*tau_x_raw*(Z==0))
s_tt1 <- sum(tau_x_raw*tau_x_raw*(Z==1))
partial_resid_mu <- resid - mu_x_raw
s_ty0 <- sum(tau_x_raw*partial_resid_mu*(Z==0))
s_ty1 <- sum(tau_x_raw*partial_resid_mu*(Z==1))
current_b_0 <- rnorm(1, (s_ty0/(s_tt0 + 2*current_sigma2)),
sqrt(current_sigma2/(s_tt0 + 2*current_sigma2)))
current_b_1 <- rnorm(1, (s_ty1/(s_tt1 + 2*current_sigma2)),
sqrt(current_sigma2/(s_tt1 + 2*current_sigma2)))
tau_basis <- (1-Z)*current_b_0 + Z*current_b_1
forest_dataset_tau$update_basis(tau_basis)
forest_model_tau$propagate_basis_update(forest_dataset_tau, outcome, active_forest_tau)
b_0_samples[i] <- current_b_0
b_1_samples[i] <- current_b_1
# Sample global variance parameter
global_var_samples[i] <- sample_sigma2_one_iteration(outcome, forest_dataset_tau, rng, nu, lambda)
current_sigma2 <- global_var_samples[i]
}
}Predict and rescale samples
# Forest predictions
mu_hat <- forest_samples_mu$predict(forest_dataset_mu)*y_std + y_bar
tau_hat_raw <- forest_samples_tau$predict_raw(forest_dataset_tau)
tau_hat <- t(t(tau_hat_raw) * (b_1_samples - b_0_samples))*y_std
y_hat <- mu_hat + tau_hat * as.numeric(Z)
# Global error variance
sigma2_samples <- global_var_samples*(y_std^2)Results
Inspect the XBART results
plot(sigma2_samples[1:num_gfr], ylab="sigma^2")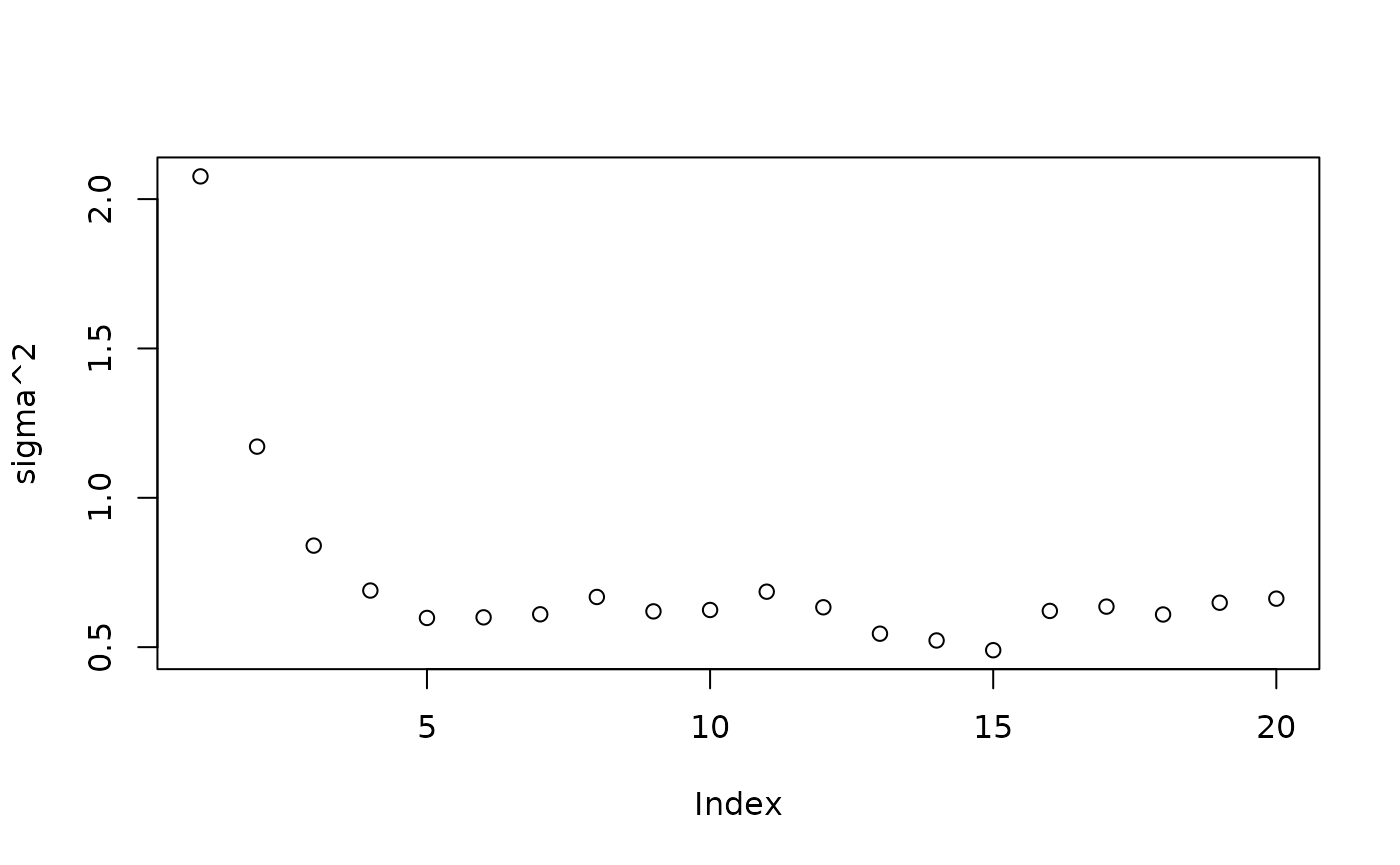
plot(rowMeans(mu_hat[,1:num_gfr]), mu_x, pch=16, cex=0.75,
xlab = "pred", ylab = "actual", main = "prognostic term")
abline(0,1,col="red",lty=2,lwd=2.5)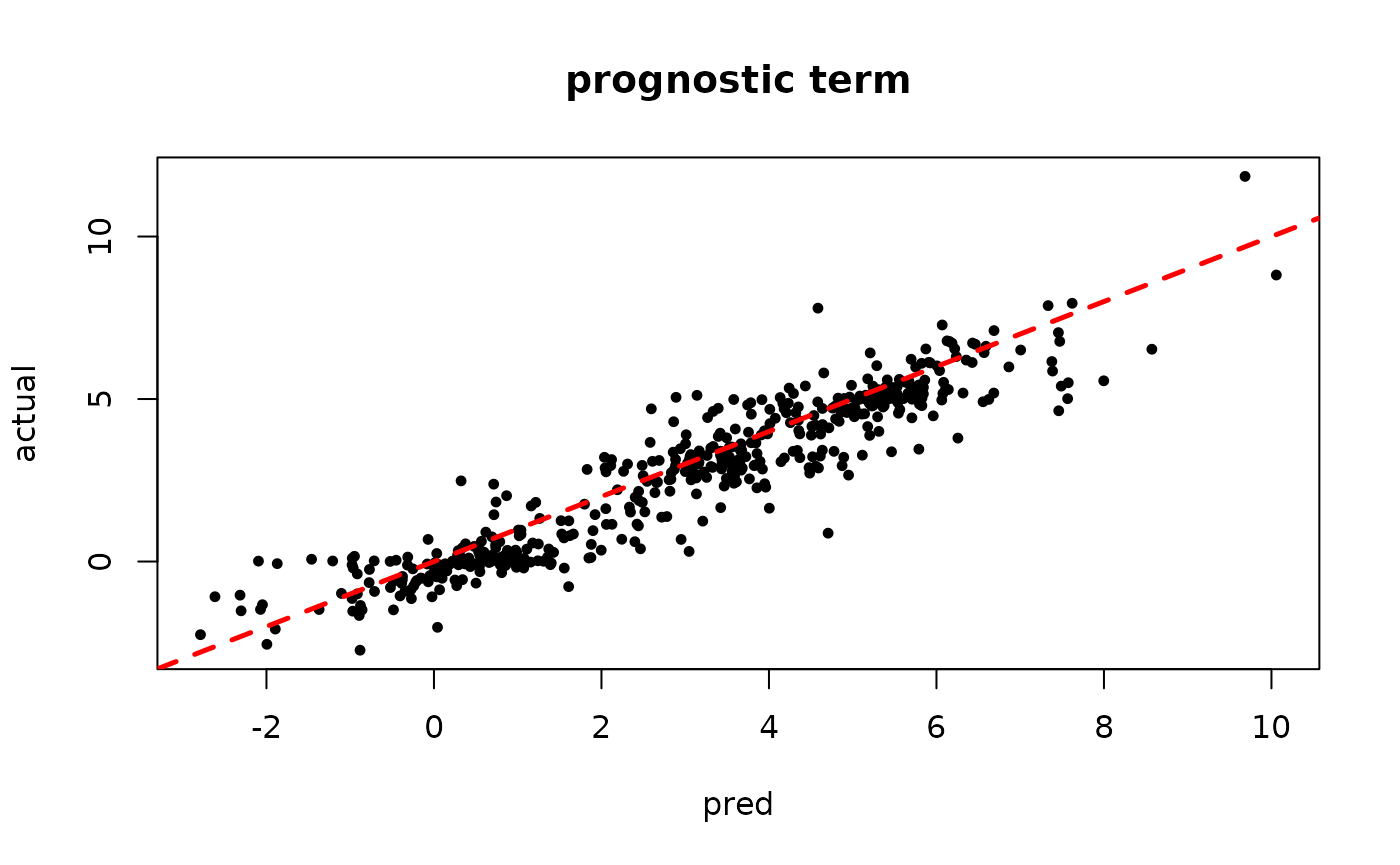
plot(rowMeans(tau_hat[,1:num_gfr]), tau_x, pch=16, cex=0.75,
xlab = "pred", ylab = "actual", main = "treatment effect term")
abline(0,1,col="red",lty=2,lwd=2.5)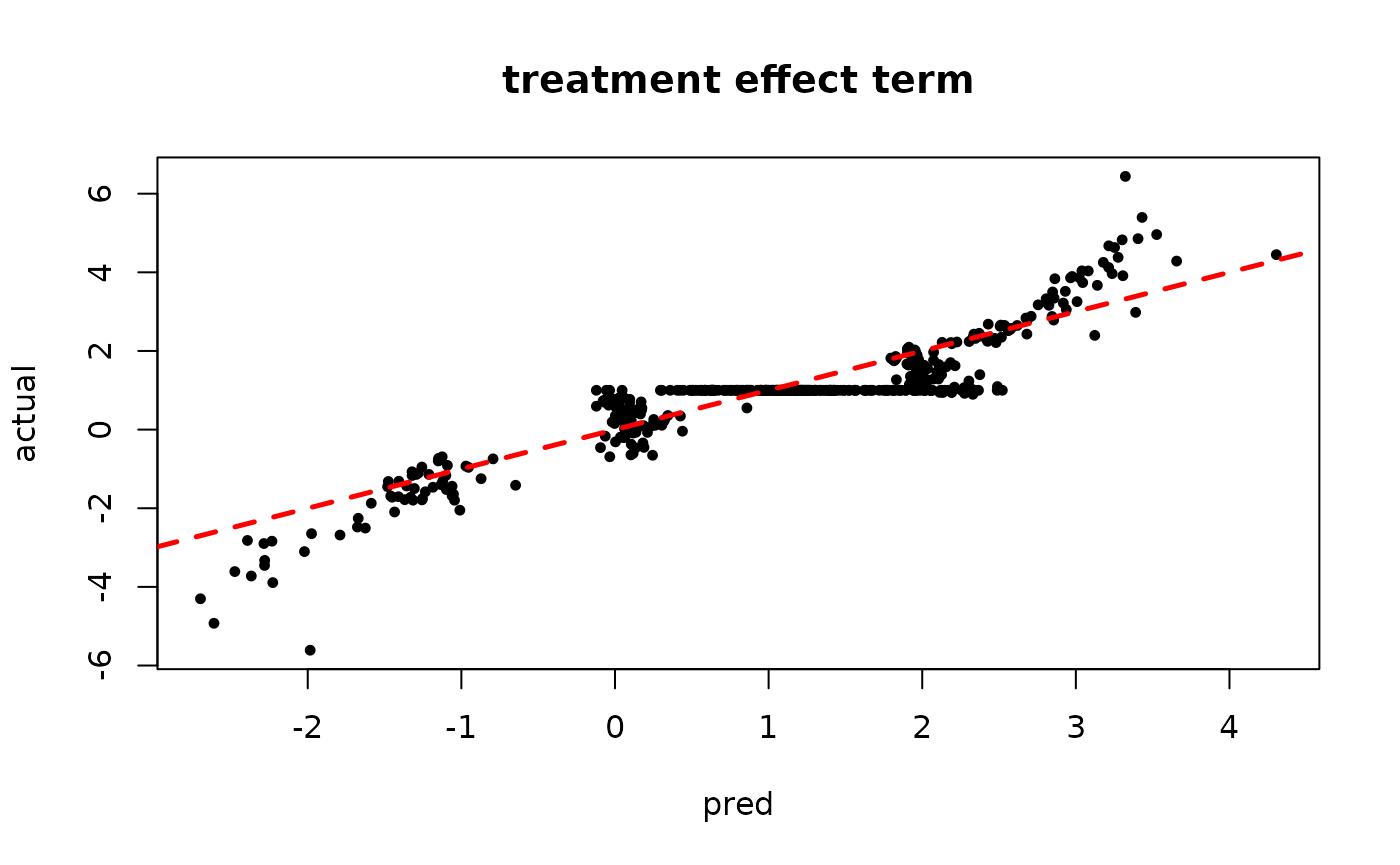
Inspect the warm start BART results
plot(sigma2_samples[(num_gfr+1):num_samples], ylab="sigma^2")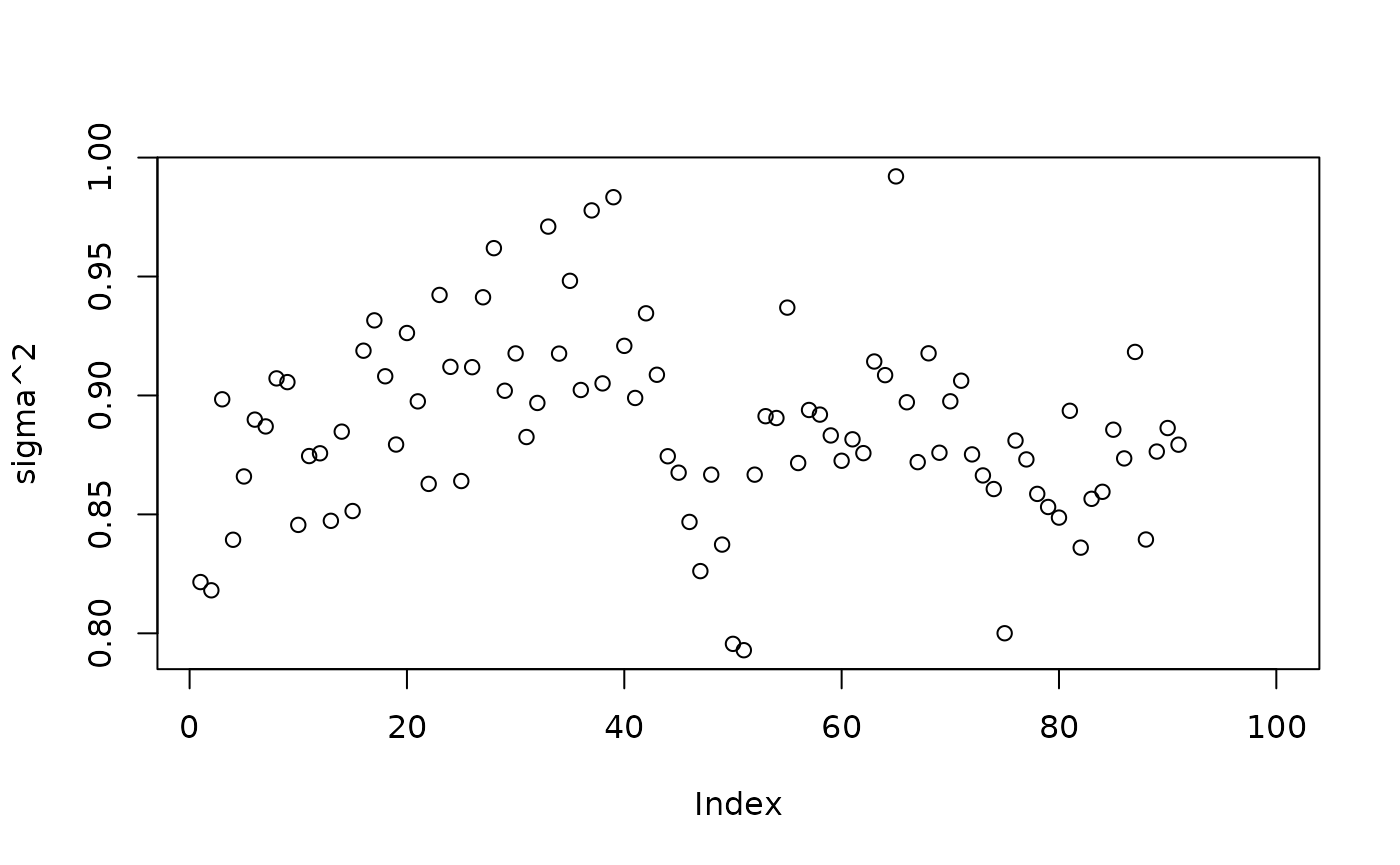
plot(rowMeans(mu_hat[,(num_gfr+1):num_samples]), mu_x, pch=16, cex=0.75,
xlab = "pred", ylab = "actual", main = "prognostic term")
abline(0,1,col="red",lty=2,lwd=2.5)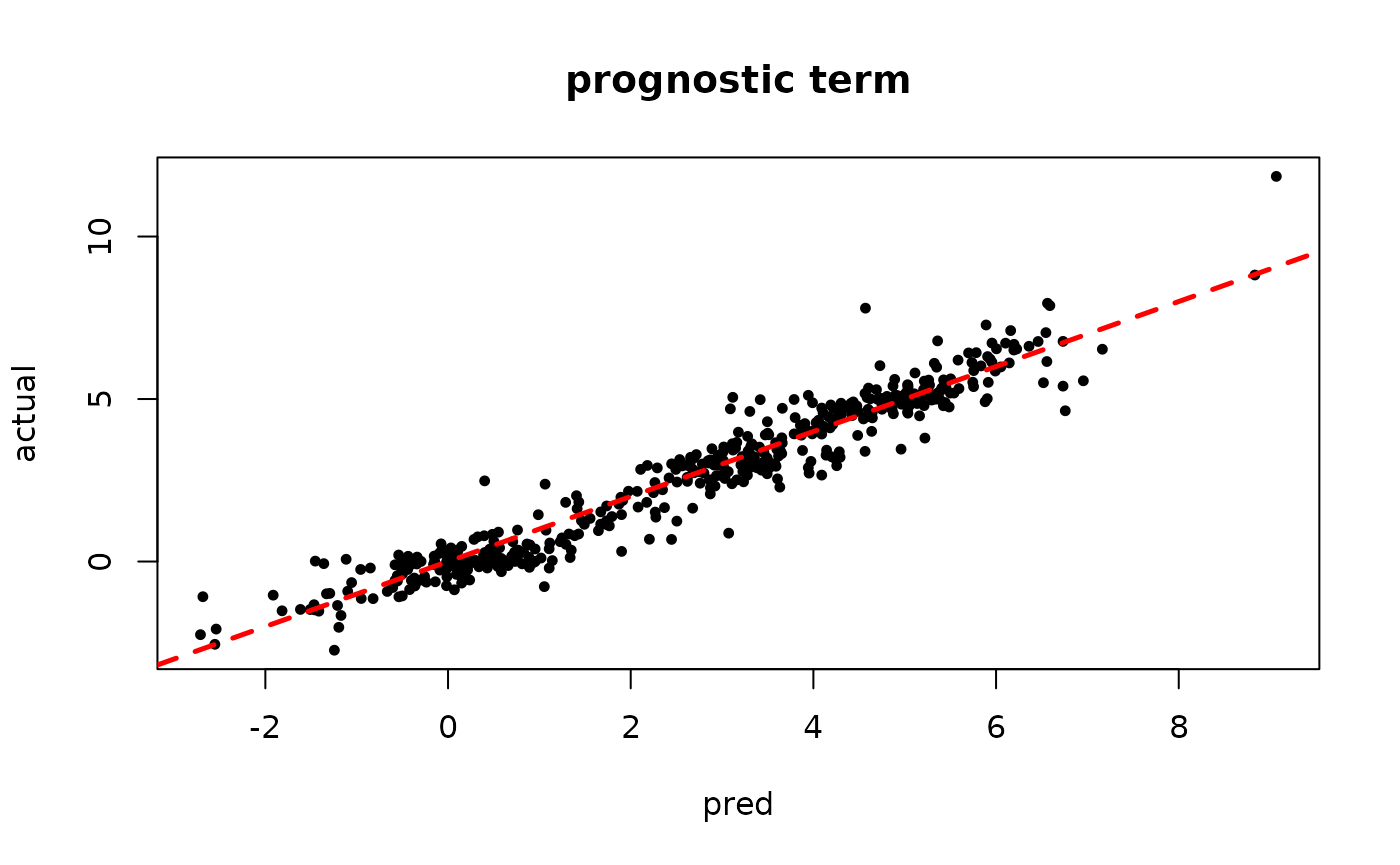
plot(rowMeans(tau_hat[,(num_gfr+1):num_samples]), tau_x, pch=16, cex=0.75,
xlab = "pred", ylab = "actual", main = "treatment effect term")
abline(0,1,col="red",lty=2,lwd=2.5)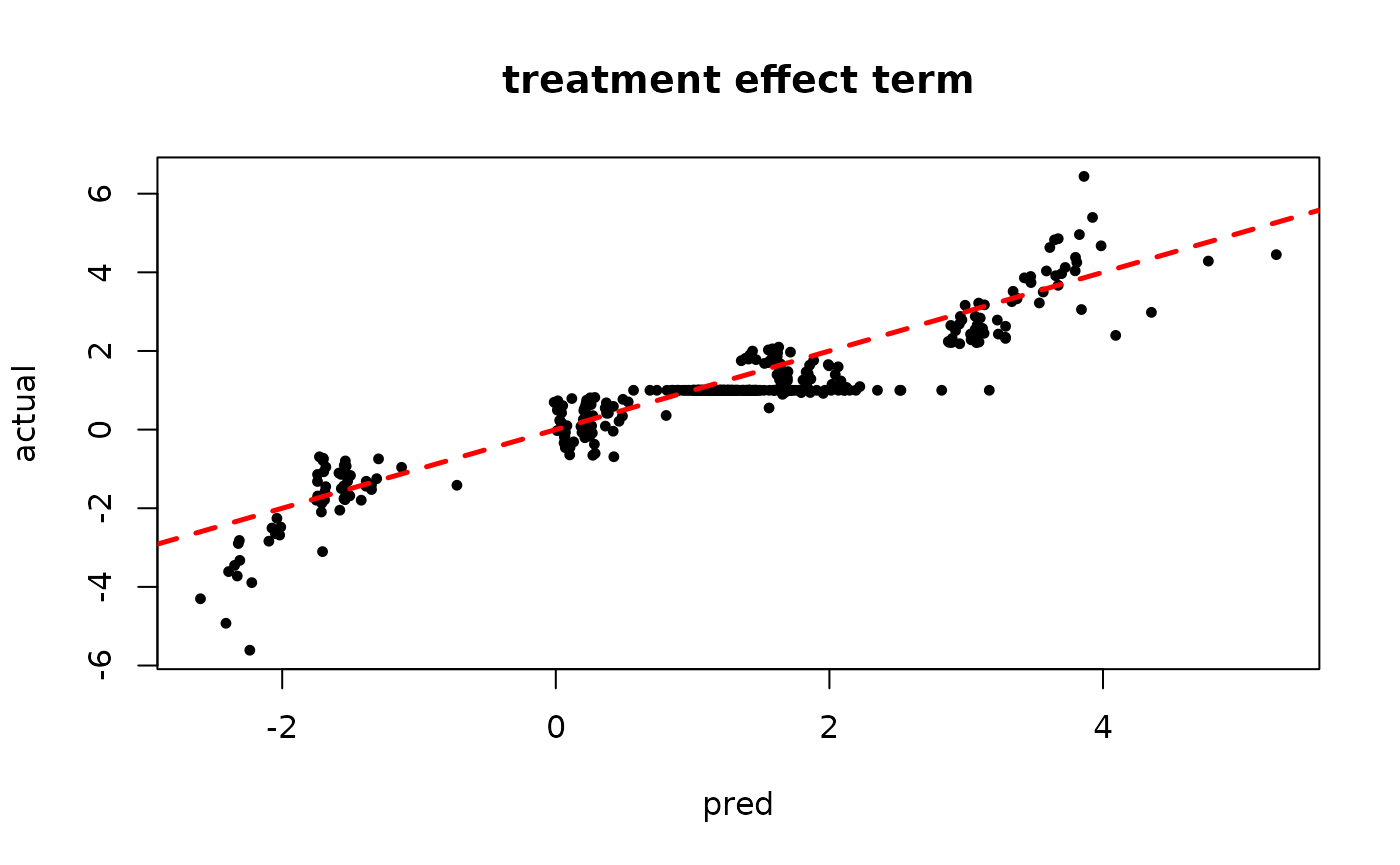
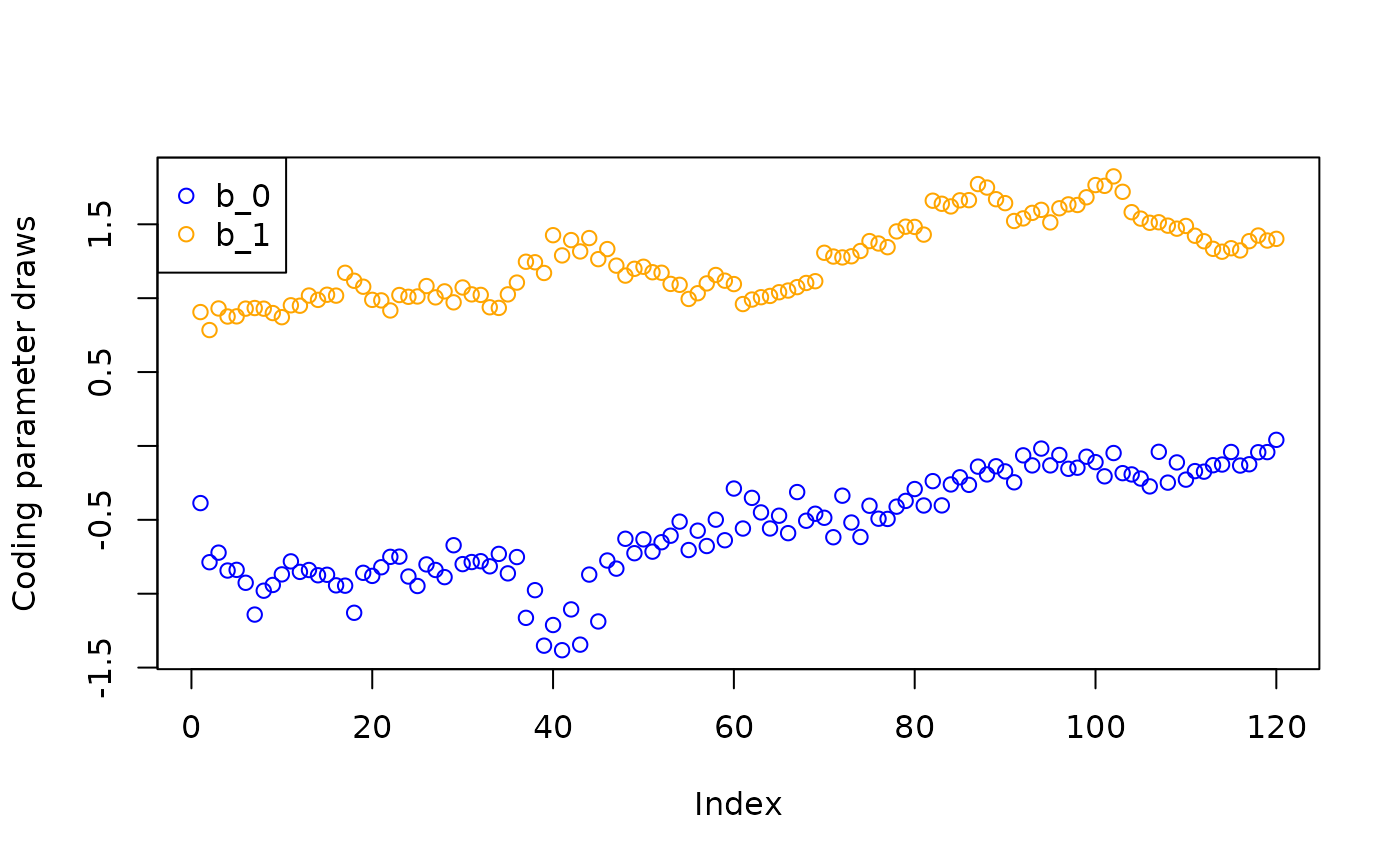
Inspect the “adaptive coding” parameters and .
plot(b_0_samples, col = "blue", ylab = "Coding parameter draws",
ylim = c(min(min(b_0_samples), min(b_1_samples)), max(max(b_0_samples), max(b_1_samples))))
points(b_1_samples, col = "orange")
legend("topleft", legend = c("b_0", "b_1"), col = c("blue", "orange"), pch = c(1,1))