Prior Calibration Approaches for Parametric Components of Stochastic Tree Ensembles
PriorCalibration.RmdBackground
The “classic” BART model of Chipman, George, and McCulloch (2010)
is semiparametric, with a nonparametric tree ensemble and a homoskedastic error variance parameter . Note that in Chipman, George, and McCulloch (2010), and are parameterized with and .
Setting Priors on Variance Parameters in stochtree
By default, stochtree employs a Jeffreys’ prior for
which corresponds to an
improper prior with
and
.
We provide convenience functions for users wishing to set the prior as in Chipman, George, and McCulloch (2010). In this case, is set by default to 3 and is calibrated as follows:
- An “overestimate,” , of is obtained via simple linear regression of on
- is chosen to ensure that for some value , typically set to a default value of 0.9.
This is done in stochtree via the
calibrate_inverse_gamma_error_variance function.
# Load library
library(stochtree)
# Generate data
n <- 500
p <- 5
X <- matrix(runif(n*p), ncol = p)
f_XW <- (
((0 <= X[,1]) & (0.25 > X[,1])) * (-7.5) +
((0.25 <= X[,1]) & (0.5 > X[,1])) * (-2.5) +
((0.5 <= X[,1]) & (0.75 > X[,1])) * (2.5) +
((0.75 <= X[,1]) & (1 > X[,1])) * (7.5)
)
noise_sd <- 1
y <- f_XW + rnorm(n, 0, noise_sd)
# Test/train split
test_set_pct <- 0.2
n_test <- round(test_set_pct*n)
n_train <- n - n_test
test_inds <- sort(sample(1:n, n_test, replace = FALSE))
train_inds <- (1:n)[!((1:n) %in% test_inds)]
X_test <- X[test_inds,]
X_train <- X[train_inds,]
y_test <- y[test_inds]
y_train <- y[train_inds]
# Calibrate the scale parameter for the variance term as in Chipman et al (2010)
nu <- 3
lambda <- calibrate_inverse_gamma_error_variance(y_train, X_train, nu = nu)Now we run a BART model with this variance parameterization
bart_params <- list(a_global = nu/2, b_global = (nu*lambda)/2)
bart_model <- bart(X_train = X_train, y_train = y_train, X_test = X_test,
num_gfr = 0, num_burnin = 1000, num_mcmc = 100,
params = bart_params)Inspect the out-of-sample predictions of the model
plot(rowMeans(bart_model$y_hat_test), y_test, xlab = "predicted", ylab = "actual")
abline(0,1,col="red",lty=3,lwd=3)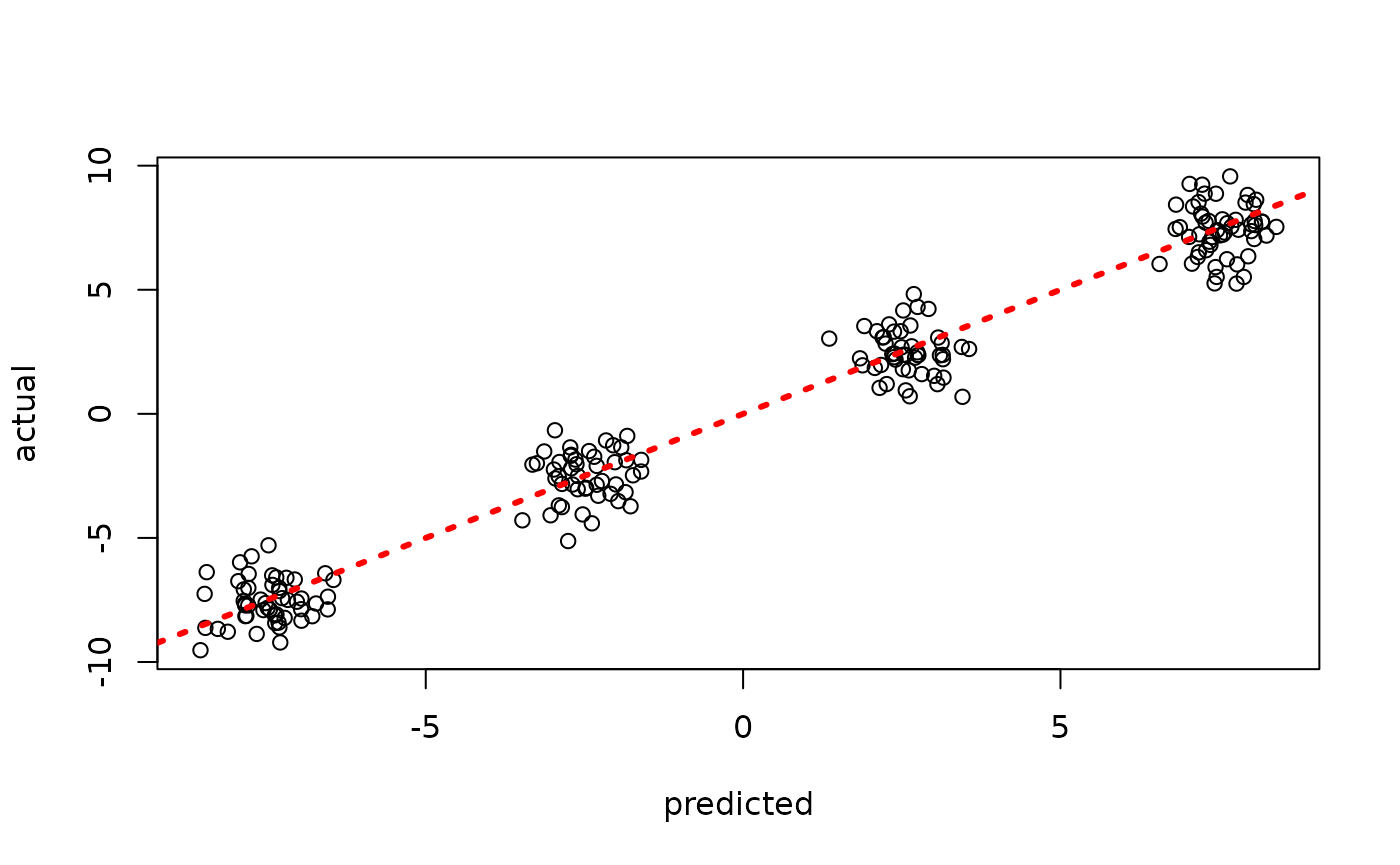
Inspect the posterior samples of
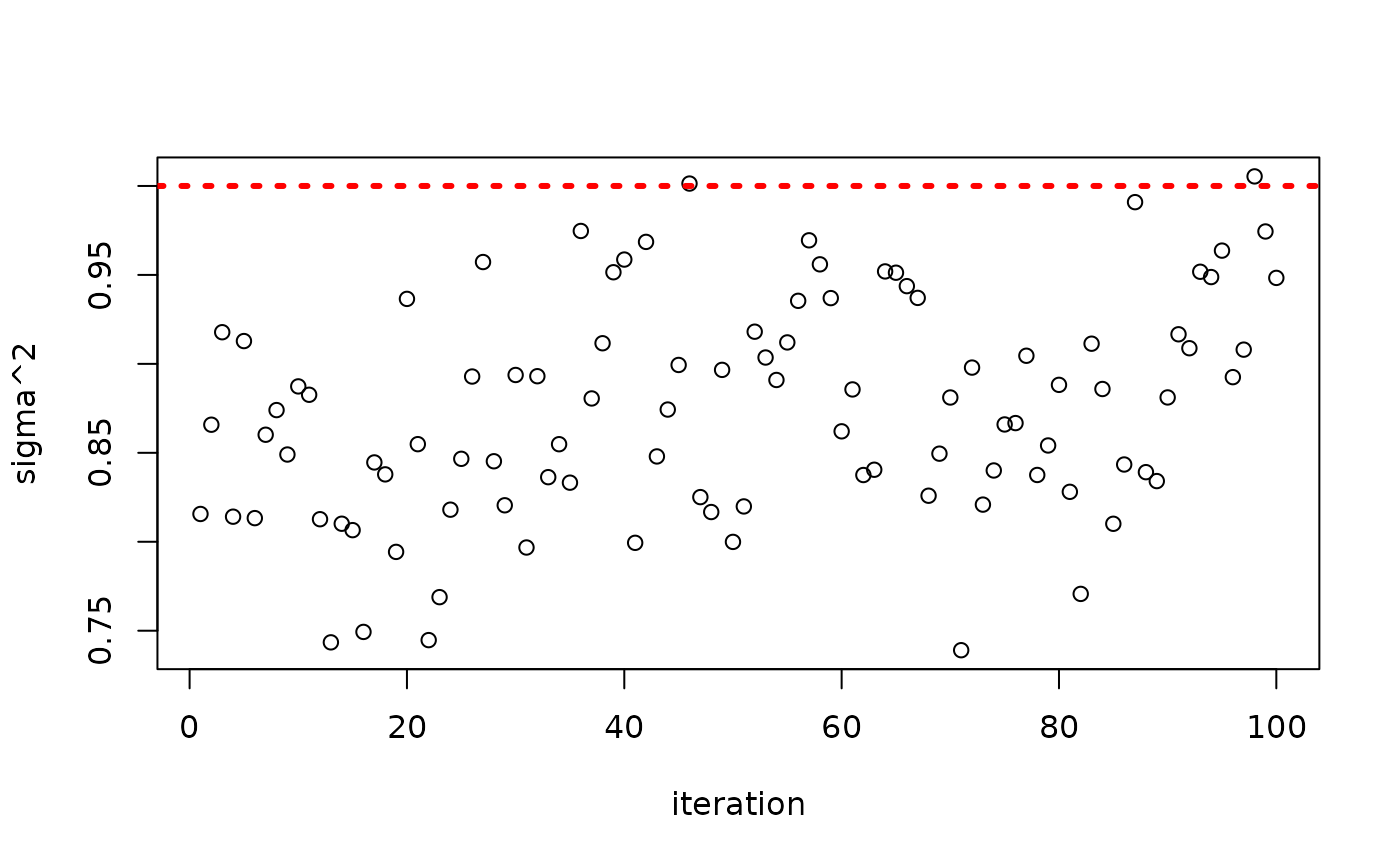
plot(bart_model$sigma2_global_samples, ylab = "sigma^2", xlab = "iteration")
abline(h = noise_sd^2, col = "red", lty = 3, lwd = 3)